Duplication of centromeric histone H3 (HTR12) gene in Arabidopsis halleri and A. lyrata, plant species with multiple centromeric satellite sequences
- PMID: 17028323
- PMCID: PMC1698631
- DOI: 10.1534/genetics.106.063628
Duplication of centromeric histone H3 (HTR12) gene in Arabidopsis halleri and A. lyrata, plant species with multiple centromeric satellite sequences
Abstract
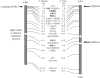
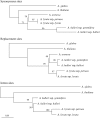
Arabidopsis halleri and lyrata have three different major centromeric satellite sequences, a unique finding for a diploid Arabidopsis species. Since centromeric histones coevolve with centromeric satellites, these proteins would be predicted to show signs of selection when new centromere satellites have recently arisen. We isolated centromeric protein genes from A. halleri and lyrata and found that one of them, HTR12 (CENP-A), is duplicated, while CENP-C is not. Phylogenetic analysis indicates that the HTR12 duplication occurred after these species diverged from A. thaliana. Genetic mapping shows that HTR12 copy B has the same genomic location as the A. thaliana gene; the other copy (A, at the other end of the same chromosome) is probably the new copy. To test for selection since the duplication, we surveyed diversity at both HTR12 loci within A. lyrata. Overall, there is no strong evidence for an "evolutionary arms race" causing multiple replacement substitutions. The A. lyrata HTR12B sequences fall into three classes of haplotypes, apparently maintained for a long time, but they all encode the same amino acid sequence. In contrast, HTR12A has low diversity, but many variants are amino acid replacements, possibly due to independent selective sweeps within populations of the species.
Figures



References
-
- Chamary, J. V., J. L. Parmley and L. D. Hurst, 2006. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 7: 98–108. - PubMed
-
- Charlesworth, B., M. Nordborg and D. Charlesworth, 1997. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided inbreeding and outcrossing populations. Genet. Res. 70: 155–174. - PubMed
-
- Cooper, J. L., and S. Henikoff, 2004. Adaptive evolution of the histone fold domain in centromeric histones. Mol. Biol. Evol. 21: 1712–1718. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

