Phylogenetic analysis of fungal centromere H3 proteins
- PMID: 17028330
- PMCID: PMC1667059
- DOI: 10.1534/genetics.106.062794
Phylogenetic analysis of fungal centromere H3 proteins
Abstract
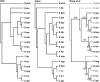
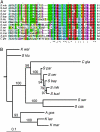
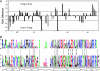
Centromere H3 proteins (CenH3's) are variants of histone H3 specialized for packaging centromere DNA. Unlike canonical H3, which is among the most conserved of eukaryotic proteins, CenH3's are rapidly evolving, raising questions about orthology and conservation of function across species. To gain insight on CenH3 evolution and function, a phylogenetic analysis was undertaken on CenH3 proteins drawn from a single, ancient lineage, the Fungi. Using maximum-likelihood methods, a credible phylogeny was derived for the conserved histone fold domain (HFD) of 25 fungal CenH3's. The collection consisted mostly of hemiascomycetous yeasts, but also included basidiomycetes, euascomycetes, and an archaeascomycete. The HFD phylogeny closely recapitulated known evolutionary relationships between the species, supporting CenH3 orthology. The fungal CenH3's lacked significant homology in their N termini except for those of the Saccharomyces/Kluyveromyces clade that all contained a region homologous to the essential N-terminal domain found in Saccharomyces cerevisiae Cse4. The ability of several heterologous CenH3's to function in S. cerevisiae was tested and found to correlate with evolutionary distance. Domain swapping between S. cerevisiae Cse4 and the noncomplementing Pichia angusta ortholog showed that species specificity could not be explained by the presence or absence of any recognized secondary structural element of the HFD.
Figures




References
-
- Berbee, M. L., and J. W. Taylor, 1993. Dating the evolutionary radiations of the true fungi. Can. J. Bot. 71: 1114–1127.
-
- Black, B. E., D. R. Foltz, S. Chakravarthy, K. Luger, V. L. Woods, Jr. et al., 2004. Structural determinants for generating centromeric chromatin. Nature 430: 578–582. - PubMed
-
- Blandin, G., B. Llorente, A. Malpertuy, P. Wincker, F. Artiguenave et al., 2000. Genomic exploration of the hemiascomycetous yeasts: 13. Pichia angusta. FEBS Lett. 487: 76–81. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

