Types and rates of sequence evolution at the high-molecular-weight glutenin locus in hexaploid wheat and its ancestral genomes
- PMID: 17028342
- PMCID: PMC1667099
- DOI: 10.1534/genetics.106.060756
Types and rates of sequence evolution at the high-molecular-weight glutenin locus in hexaploid wheat and its ancestral genomes
Abstract
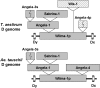
The Glu-1 locus, encoding the high-molecular-weight glutenin protein subunits, controls bread-making quality in hexaploid wheat (Triticum aestivum) and represents a recently evolved region unique to Triticeae genomes. To understand the molecular evolution of this locus region, three orthologous Glu-1 regions from the three subgenomes of a single hexaploid wheat species were sequenced, totaling 729 kb of sequence. Comparing each Glu-1 region with its corresponding homologous region from the D genome of diploid wheat, Aegilops tauschii, and the A and B genomes of tetraploid wheat, Triticum turgidum, revealed that, in addition to the conservation of microsynteny in the genic regions, sequences in the intergenic regions, composed of blocks of nested retroelements, are also generally conserved, although a few nonshared retroelements that differentiate the homologous Glu-1 regions were detected in each pair of the A and D genomes. Analysis of the indel frequency and the rate of nucleotide substitution, which represent the most frequent types of sequence changes in the Glu-1 regions, demonstrated that the two A genomes are significantly more divergent than the two B genomes, further supporting the hypothesis that hexaploid wheat may have more than one tetraploid ancestor.
Figures



References
-
- Allaby, R. G., M. Banerjee and T. A. Brown, 1999. Evolution of the high molecular weight glutenin loci of the A, B, D, and G genomes of wheat. Genome 42: 296–307. - PubMed
-
- Anderson, O. D., C. Rausch, O. Moullet and E. S. Lagudah, 2003. The wheat D-genome HMW-glutenin locus: BAC sequencing, gene distribution, and retrotransposon cluster. Funct. Integr. Genomics 3: 56–68. - PubMed
-
- Blake, N. K., B. R. Lehfeldt, M. Lavin and L. E. Talbert, 1999. Phylogenetic reconstruction based on low copy DNA sequence data in an allopolyploid: the B genome of wheat. Genome 42: 351–360. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

