Natural biocombinatorics in the polyketide synthase genes of the actinobacterium Streptomyces avermitilis
- PMID: 17029557
- PMCID: PMC1592344
- DOI: 10.1371/journal.pcbi.0020132
Natural biocombinatorics in the polyketide synthase genes of the actinobacterium Streptomyces avermitilis
Abstract
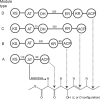
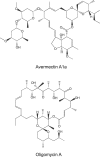
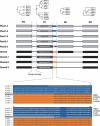
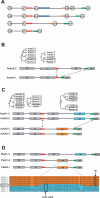
Modular polyketide synthases (PKSs) of bacteria provide an enormous reservoir of natural chemical diversity. Studying natural biocombinatorics may aid in the development of concepts for experimental design of genes for the biosynthesis of new bioactive compounds. Here we address the question of how the modularity of biosynthetic enzymes and the prevalence of multiple gene clusters in Streptomyces drive the evolution of metabolic diversity. The phylogeny of ketosynthase (KS) domains of Streptomyces PKSs revealed that the majority of modules involved in the biosynthesis of a single compound evolved by duplication of a single ancestor module. Using Streptomyces avermitilis as a model organism, we have reconstructed the evolutionary relationships of different domain types. This analysis suggests that 65% of the modules were altered by recombinational replacements that occurred within and between biosynthetic gene clusters. The natural reprogramming of the biosynthetic pathways was unambiguously confined to domains that account for the structural diversity of the polyketide products and never observed for the KS domains. We provide examples for natural acyltransferase (AT), ketoreductase (KR), and dehydratase (DH)-KR domain replacements. Potential sites of homologous recombination could be identified in interdomain regions and within domains. Our results indicate that homologous recombination facilitated by the modularity of PKS architecture is the most important mechanism underlying polyketide diversity in bacteria.
Conflict of interest statement
Figures
References
-
- Staunton J, Weissman KJ. Polyketide biosynthesis: A millennium review. Nat Prod Rep. 2001;18:380–416. - PubMed
-
- Hopwood DA, Sherman DH. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu Rev Genet. 1990;24:37–66. - PubMed
-
- Caffrey P. Conserved amino acid residues correlating with ketoreductase stereospecificity in modular polyketide synthases. Chembiochem. 2003;4:654–657. - PubMed
-
- Gonzalez-Lergier J, Broadbelt LJ, Hatzimanikatis V. Theoretical considerations and computational analysis of the complexity in polyketide synthesis pathways. J Am Chem Soc. 2005;127:9930–9938. - PubMed
-
- Hopwood DA. Genetic contributions to understanding polyketide synthases. Chem Rev. 1997;97:2465–2498. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

