Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase zeta for mismatch extension and for extension opposite from DNA lesions
- PMID: 17030609
- PMCID: PMC1698531
- DOI: 10.1128/MCB.01671-06
Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase zeta for mismatch extension and for extension opposite from DNA lesions
Abstract
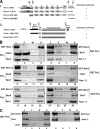
Rev1, a Y family DNA polymerase (Pol) functions together with Polzeta, a B family Pol comprised of the Rev3 catalytic subunit and Rev7 accessory subunit, in promoting translesion DNA synthesis (TLS). Extensive genetic studies with Saccharomyces cerevisiae have indicated a requirement of both Polzeta and Rev1 for damage-induced mutagenesis, implicating their involvement in mutagenic TLS. Polzeta is specifically adapted to promote the extension step of lesion bypass, as it proficiently extends primer termini opposite DNA lesions, and it is also a proficient extender of mismatched primer termini on undamaged DNAs. Since TLS through UV-induced lesions and various other DNA lesions does not depend upon the DNA-synthetic activity of Rev1, Rev1 must contribute to Polzeta-dependent TLS in a nonenzymatic way. Here, we provide evidence for the physical association of Rev1 with Polzeta and show that this binding is mediated through the C terminus of Rev1 and the polymerase domain of Rev3. Importantly, a rev1 mutant that lacks the C-terminal 72 residues which inactivate interaction with Rev3 exhibits the same high degree of UV sensitivity and defectiveness in UV-induced mutagenesis as that conferred by the rev1Delta mutation. We propose that Rev1 binding to Polzeta is indispensable for the targeting of Polzeta to the replication fork stalled at a DNA lesion. In addition to this structural role, Rev1 binding enhances the proficiency of Polzeta for the extension of mismatched primer termini on undamaged DNAs and for the extension of primer termini opposite DNA lesions.
Figures


References
-
- Bailly, V., J. Lamb, P. Sung, S. Prakash, and L. Prakash. 1994. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 8:811-820. - PubMed
-
- Bailly, V., S. Lauder, S. Prakash, and L. Prakash. 1997. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 272:23360-23365. - PubMed
-
- Baynton, K., A. Bresson-Roy, and R. P. P. Fuchs. 1999. Distinct roles for Rev1p and Rev7p during translesion synthesis in Saccharomyces cerevisiae. Mol. Microbiol. 34:124-133. - PubMed
-
- Gerik, K. J., X. Li, A. Pautz, and P. M. J. Burgers. 1998. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 273:19747-19755. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
