Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility
- PMID: 17030797
- PMCID: PMC1622818
- DOI: 10.1073/pnas.0606924103
Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility
Abstract
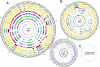
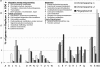
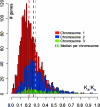
Burkholderia xenovorans LB400 (LB400), a well studied, effective polychlorinated biphenyl-degrader, has one of the two largest known bacterial genomes and is the first nonpathogenic Burkholderia isolate sequenced. From an evolutionary perspective, we find significant differences in functional specialization between the three replicons of LB400, as well as a more relaxed selective pressure for genes located on the two smaller vs. the largest replicon. High genomic plasticity, diversity, and specialization within the Burkholderia genus are exemplified by the conservation of only 44% of the genes between LB400 and Burkholderia cepacia complex strain 383. Even among four B. xenovorans strains, genome size varies from 7.4 to 9.73 Mbp. The latter is largely explained by our findings that >20% of the LB400 sequence was recently acquired by means of lateral gene transfer. Although a range of genetic factors associated with in vivo survival and intercellular interactions are present, these genetic factors are likely related to niche breadth rather than determinants of pathogenicity. The presence of at least eleven "central aromatic" and twenty "peripheral aromatic" pathways in LB400, among the highest in any sequenced bacterial genome, supports this hypothesis. Finally, in addition to the experimentally observed redundancy in benzoate degradation and formaldehyde oxidation pathways, the fact that 17.6% of proteins have a better LB400 paralog than an ortholog in a different genome highlights the importance of gene duplication and repeated acquirement, which, coupled with their divergence, raises questions regarding the role of paralogs and potential functional redundancies in large-genome microbes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

