Proton-proton Overhauser NMR spectroscopy with polypeptide chains in large structures
- PMID: 17032756
- PMCID: PMC1622842
- DOI: 10.1073/pnas.0607141103
Proton-proton Overhauser NMR spectroscopy with polypeptide chains in large structures
Abstract
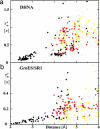
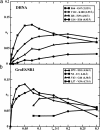
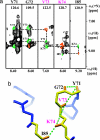
The use of 1H-1H nuclear Overhauser effects (NOE) for structural studies of uniformly deuterated polypeptide chains in large structures is investigated by model calculations and NMR experiments. Detailed analysis of the evolution of the magnetization during 1H-1H NOE experiments under slow-motion conditions shows that the maximal 1H-1H NOE transfer is independent of the overall rotational correlation time, even in the presence of chemical exchange with the bulk water, provided that the mixing time is adjusted for the size of the structure studied. 1H-1H NOE buildup measurements were performed for the 472-kDa complex of the 72-kDa cochaperonin GroES with a 400-kDa single-ring variant of the chaperonin GroEL (SR1). These experiments demonstrate that multidimensional NOESY experiments with cross-correlated relaxation-enhanced polarization transfer and transverse relaxation-optimized spectroscopy elements can be applied to structures of molecular masses up to several hundred kilodaltabs, which opens new possibilities for studying functional interactions in large maromolecular assemblies in solution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

