Nucleotide-resolution mapping of topoisomerase-mediated and apoptotic DNA strand scissions at or near an MLL translocation hotspot
- PMID: 17033956
- PMCID: PMC1698565
- DOI: 10.1086/507791
Nucleotide-resolution mapping of topoisomerase-mediated and apoptotic DNA strand scissions at or near an MLL translocation hotspot
Abstract
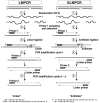
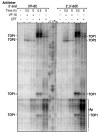
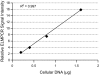
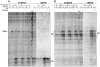
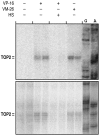
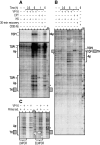
The emergence of therapy-related acute myeloid leukemia (t-AML) has been associated with DNA topoisomerase II (TOP2)-targeted drug treatments and chromosomal translocations frequently involving the MLL, or ALL-1, gene. Two distinct mechanisms have been implicated as potential triggers of t-AML translocations: TOP2-mediated DNA cleavage and apoptotic higher-order chromatin fragmentation. Assessment of the role of TOP2 in this process has been hampered by a lack of techniques allowing in vivo mapping of TOP2-mediated DNA cleavage at nucleotide resolution in single-copy genes. A novel method, extension ligation-mediated polymerase chain reaction (ELMPCR), was used here for mapping topoisomerase-mediated DNA strand breaks and apoptotic DNA cleavage across a translocation-prone region of MLL in human cells. We report the first genomic map integrating translocation breakpoints and topoisomerase I, TOP2, and apoptotic DNA cleavage sites at nucleotide resolution across an MLL region harboring a t-AML translocation hotspot. This hotspot is flanked by a TOP2 cleavage site and is localized at one extremity of a minor apoptotic cleavage region, where multiple single- and double-strand breaks were induced by caspase-activated apoptotic nucleases. This cleavage pattern was in sharp contrast to that observed approximately 200 bp downstream in the exon 12 region, which displayed much stronger apoptotic cleavage but where no double-strand breaks were detected and no t-AML-associated breakpoints were reported. The localization and remarkable clustering of the t-AML breakpoints cannot be explained simply by the DNA cleavage patterns but might result from potential interactions between TOP2 poisoning, apoptotic DNA cleavage, and DNA repair attempts at specific sites of higher-order chromatin structure in apoptosis-evading cells. ELMPCR provides a new tool for investigating the role of DNA topoisomerases in fundamental genetic processes and translocations associated with cancer treatments involving topoisomerase-targeted drugs.
Figures


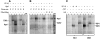

References
Web Resources
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for MLL sequence [accession number U04737])
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MLL/ALL-1) - PubMed
References
-
- Pedersen-Bjergaard J, Rowley JD (1994) The balanced and the unbalanced chromosome aberrations of acute myeloid leukemia may develop in different ways and may contribute differently to malignant transformation. Blood 83:2780–2786 - PubMed
-
- Felix CA (1998) Secondary leukemias induced by topoisomerase-targeted drugs. Biochim Biophys Acta 1400:233–255 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

