Test for interaction between two unlinked loci
- PMID: 17033960
- PMCID: PMC1698572
- DOI: 10.1086/508571
Test for interaction between two unlinked loci
Abstract
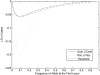
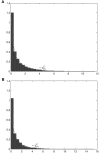
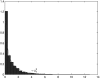
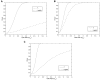
Despite the growing consensus on the importance of testing gene-gene interactions in genetic studies of complex diseases, the effect of gene-gene interactions has often been defined as a deviance from genetic additive effects, which is essentially treated as a residual term in genetic analysis and leads to low power in detecting the presence of interacting effects. To what extent the definition of gene-gene interaction at population level reflects the genes' biochemical or physiological interaction remains a mystery. In this article, we introduce a novel definition and a new measure of gene-gene interaction between two unlinked loci (or genes). We developed a general theory for studying linkage disequilibrium (LD) patterns in disease population under two-locus disease models. The properties of using the LD measure in a disease population as a function of the measure of gene-gene interaction between two unlinked loci were also investigated. We examined how interaction between two loci creates LD in a disease population and showed that the mathematical formulation of the new definition for gene-gene interaction between two loci was similar to that of the LD between two loci. This finding motived us to develop an LD-based statistic to detect gene-gene interaction between two unlinked loci. The null distribution and type I error rates of the LD-based statistic for testing gene-gene interaction were validated using extensive simulation studies. We found that the new test statistic was more powerful than the traditional logistic regression under three two-locus disease models and demonstrated that the power of the test statistic depends on the measure of gene-gene interaction. We also investigated the impact of using tagging SNPs for testing interaction on the power to detect interaction between two unlinked loci. Finally, to evaluate the performance of our new method, we applied the LD-based statistic to two published data sets. Our results showed that the P values of the LD-based statistic were smaller than those obtained by other approaches, including logistic regression models.
Figures



References
-
- Fisher RA (1918) The correlation between relatives on the supposition of Mendelian inheritance. Trans R Soc Edinb 3:399–433
-
- Kempthorne O (1954) The correlation between relatives in a random mating population. Proc R Soc Lond B 143:103–113 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

