Ubiquitin-binding domains
- PMID: 17034365
- PMCID: PMC1615911
- DOI: 10.1042/BJ20061138
Ubiquitin-binding domains
Abstract
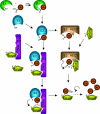
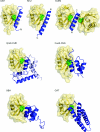
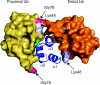
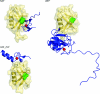
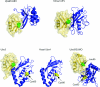
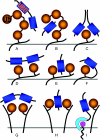
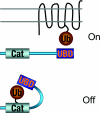
The covalent modification of proteins by ubiquitination is a major regulatory mechanism of protein degradation and quality control, endocytosis, vesicular trafficking, cell-cycle control, stress response, DNA repair, growth-factor signalling, transcription, gene silencing and other areas of biology. A class of specific ubiquitin-binding domains mediates most of the effects of protein ubiquitination. The known membership of this group has expanded rapidly and now includes at least sixteen domains: UBA, UIM, MIU, DUIM, CUE, GAT, NZF, A20 ZnF, UBP ZnF, UBZ, Ubc, UEV, UBM, GLUE, Jab1/MPN and PFU. The structures of many of the complexes with mono-ubiquitin have been determined, revealing interactions with multiple surfaces on ubiquitin. Inroads into understanding polyubiquitin specificity have been made for two UBA domains, whose structures have been characterized in complex with Lys48-linked di-ubiquitin. Several ubiquitin-binding domains, including the UIM, CUE and A20 ZnF (zinc finger) domains, promote auto-ubiquitination, which regulates the activity of proteins that contain them. At least one of these domains, the A20 ZnF, acts as a ubiquitin ligase by recruiting a ubiquitin-ubiquitin-conjugating enzyme thiolester adduct in a process that depends on the ubiquitin-binding activity of the A20 ZnF. The affinities of the mono-ubiquitin-binding interactions of these domains span a wide range, but are most commonly weak, with Kd>100 microM. The weak interactions between individual domains and mono-ubiquitin are leveraged into physiologically relevant high-affinity interactions via several mechanisms: ubiquitin polymerization, modification multiplicity, oligomerization of ubiquitinated proteins and binding domain proteins, tandem-binding domains, binding domains with multiple ubiquitin-binding sites and co-operativity between ubiquitin binding and binding through other domains to phospholipids and small G-proteins.
Figures
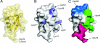
References
-
- Hershko A., Ciechanover A., Varshavsky A. The ubiquitin system. Nat. Med. 2000;6:1073–1081. - PubMed
-
- Pickart C. M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. - PubMed
-
- Weissman A. M. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2001;2:169–178. - PubMed
-
- Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 2000;2:E153–E157. - PubMed
-
- Miller J., Gordon C. The regulation of proteasome degradation by multi-ubiquitin chain binding proteins. FEBS Lett. 2005;579:3224–3230. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

