A novel analytical method for in vivo phosphate tracking
- PMID: 17034793
- PMCID: PMC2748124
- DOI: 10.1016/j.febslet.2006.09.048
A novel analytical method for in vivo phosphate tracking
Erratum in
- FEBS Lett. 2007 Feb 6;581(3):579
Abstract
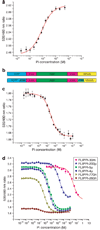
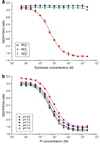
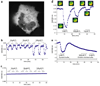
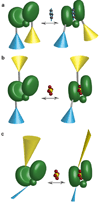
Genetically-encoded fluorescence resonance energy transfer (FRET) sensors for phosphate (P(i)) (FLIPPi) were engineered by fusing a predicted Synechococcus phosphate-binding protein (PiBP) to eCFP and Venus. Purified fluorescent indicator protein for inorganic phosphate (FLIPPi), in which the fluorophores are attached to the same PiBP lobe, shows P(i)-dependent increases in FRET efficiency. FLIPPi affinity mutants cover P(i) changes over eight orders of magnitude. COS-7 cells co-expressing a low-affinity FLIPPi and a Na(+)/P(i) co-transporter exhibited FRET changes when perfused with 100 microM P(i), demonstrating concentrative P(i) uptake by PiT2. FLIPPi sensors are suitable for real-time monitoring of P(i) metabolism in living cells, providing a new tool for fluxomics, analysis of pathophysiology or changes of P(i) during cell migration.
Figures

References
-
- Collins JF, Bai L, Ghishan FK. The SLC20 family of proteins: dual functions as sodium-phosphate cotransporters and viral receptors. Pflügers Arch. 2004;447:647–652. - PubMed
-
- Vidal G, Gallis JL, Dufour S, Canioni P. NMR studies of inorganic phosphate compartmentation in the isolated rat liver during acidic perfusion. Arch. Biochem. Biophys. 1997;337:317–325. - PubMed
-
- Wang ZM, Choudhary A, Ledvina PS, Quiocho FA. Fine-tuning the specificity of the periplasmic phosphate transport receptor – site-directed mutagenesis, ligand-binding, and crystallographic studies. J. Biol. Chem. 1994;269:25091–25094. - PubMed
-
- Medveczky N, Rosenberg H. Phosphate-binding protein of Escherichia coli. Biochim. Biophys. Acta. 1970;211:158.
-
- Brune M, Hunter JL, Corrie JET, Webb MR. Direct, real-time measurement of rapid inorganic-phosphate release using a novel fluorescent probe and its application to actomyosin subfragment-1 ATPase. Biochemistry. 1994;33:8262–8271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

