Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD
- PMID: 17035313
- PMCID: PMC1797233
- DOI: 10.1128/JVI.01842-06
Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD
Abstract
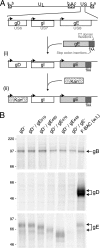
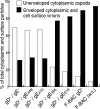
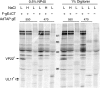
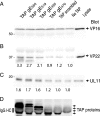
The final assembly of herpes simplex virus (HSV) involves binding of tegument-coated capsids to viral glycoprotein-enriched regions of the trans-Golgi network (TGN) as enveloped virions bud into TGN membranes. We previously demonstrated that HSV glycoproteins gE/gI and gD, acting in a redundant fashion, are essential for this secondary envelopment. To define regions of the cytoplasmic (CT) domain of gE required for secondary envelopment, HSVs lacking gD and expressing truncated gE molecules were constructed. A central region (amino acids 470 to 495) of the gE CT domain was important for secondary envelopment, although more C-terminal residues also contributed. Tandem affinity purification (TAP) proteins including fragments of the gE CT domain were used to identify tegument proteins VP22 and UL11 as binding partners, and gE CT residues 470 to 495 were important in this binding. VP22 and UL11 were precipitated from HSV-infected cells in conjunction with full-length gE and gE molecules with more-C-terminal residues of the CT domain. gD also bound VP22 and UL11. Expression of VP22 and gD or gE/gI in cells by use of adenovirus (Ad) vectors provided evidence that other viral proteins were not necessary for tegument/glycoprotein interactions. Substantial quantities of VP22 and UL11 bound nonspecifically onto or were precipitated with gE and gD molecules lacking all CT sequences, something that is very unlikely in vivo. VP16 was precipitated equally whether gE/gI or gD was present in extracts or not. These observations illustrated important properties of tegument proteins. VP22, UL11, and VP16 are highly prone to binding nonspecifically to other proteins, and this did not represent insolubility during our assays. Rather, it likely reflects an inherent "stickiness" related to the formation of tegument. Nevertheless, assays involving TAP proteins and viral proteins expressed by HSV and Ad vectors supported the conclusion that VP22 and UL11 interact specifically with the CT domains of gD and gE.
Figures





References
-
- Ball, J. M., Z. Moldoveanu, L. R. Melsen, P. A. Kozlowski, S. Jackson, M. J. Mulligan, J. F. Mestecky, and R. W. Compans. 1995. A polarized human endometrial cell line that binds and transports polymeric IgA. In Vitro Cell. Dev. Biol. Anim. 31:196-206. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

