Evolution of competence and DNA uptake specificity in the Pasteurellaceae
- PMID: 17038178
- PMCID: PMC1626085
- DOI: 10.1186/1471-2148-6-82
Evolution of competence and DNA uptake specificity in the Pasteurellaceae
Abstract
Background: Many bacteria can take up DNA, but the evolutionary history and function of natural competence and transformation remain obscure. The sporadic distribution of competence suggests it is frequently lost and/or gained, but this has not been examined in an explicitly phylogenetic context. Additional insight may come from the sequence specificity of uptake by species such as Haemophilus influenzae, where a 9 bp uptake signal sequence (USS) repeat is both highly overrepresented in the genome and needed for efficient DNA uptake. We used the distribution of competence genes and DNA uptake specificity in H. influenzae's family, the Pasteurellaceae, to examine the ancestry of competence.
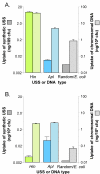
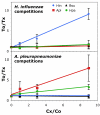
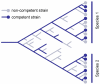
Results: A phylogeny of the Pasteurellaceae based on 12 protein coding genes from species with sequenced genomes shows two strongly supported subclades: the Hin subclade (H. influenzae, Actinobacillus actinomycetemcomitans, Pasteurella multocida, Mannheimia succiniciproducens, and H. somnus), and the Apl subclade (A. pleuropneumoniae, M. haemolytica, and H. ducreyi). All species contained homologues of all known H. influenzae competence genes, consistent with an ancestral origin of competence. Competence gene defects were identified in three species (H. somnus, H. ducreyi and M. haemolytica); each appeared to be of recent origin. The assumption that USS arise by mutation rather than copying was first confirmed using alignments of H. influenzae proteins with distant homologues. Abundant USS-like repeats were found in all eight Pasteurellacean genomes; the repeat consensuses of species in the Hin subclade were identical to that of H. influenzae (AAGTGCGGT), whereas members of the Apl subclade shared the consensus ACAAGCGGT. All species' USSs had the strong consensus and flanking AT-rich repeats of H. influenzae USSs. DNA uptake and competition experiments demonstrated that the Apl-type repeat is a true USS distinct from the Hin-type USS: A. pleuropneumoniae preferentially takes up DNA fragments containing the Apl-type USS over both H. influenzae and unrelated DNAs, and H. influenzae prefers its own USS over the Apl type.
Conclusion: Competence and DNA uptake specificity are ancestral properties of the Pasteurellaceae, with divergent USSs and uptake specificity distinguishing only the two major subclades. The conservation of most competence genes over the approximately 350 million year history of the family suggests that lineages that lose competence may be evolutionary dead ends.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

