Integration of curated databases to identify genotype-phenotype associations
- PMID: 17038185
- PMCID: PMC1630430
- DOI: 10.1186/1471-2164-7-257
Integration of curated databases to identify genotype-phenotype associations
Abstract
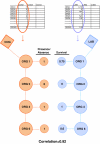
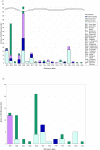
Background: The ability to rapidly characterize an unknown microorganism is critical in both responding to infectious disease and biodefense. To do this, we need some way of anticipating an organism's phenotype based on the molecules encoded by its genome. However, the link between molecular composition (i.e. genotype) and phenotype for microbes is not obvious. While there have been several studies that address this challenge, none have yet proposed a large-scale method integrating curated biological information. Here we utilize a systematic approach to discover genotype-phenotype associations that combines phenotypic information from a biomedical informatics database, GIDEON, with the molecular information contained in National Center for Biotechnology Information's Clusters of Orthologous Groups database (NCBI COGs).
Results: Integrating the information in the two databases, we are able to correlate the presence or absence of a given protein in a microbe with its phenotype as measured by certain morphological characteristics or survival in a particular growth media. With a 0.8 correlation score threshold, 66% of the associations found were confirmed by the literature and at a 0.9 correlation threshold, 86% were positively verified.
Conclusion: Our results suggest possible phenotypic manifestations for proteins biochemically associated with sugar metabolism and electron transport. Moreover, we believe our approach can be extended to linking pathogenic phenotypes with functionally related proteins.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

