Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial
- PMID: 17038190
- PMCID: PMC1794565
- DOI: 10.1186/gb-2006-7-10-r90
Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial
Abstract
Background: Pseudomonas aeruginosa is a ubiquitous environmental bacterium and an important opportunistic human pathogen. Generally, the acquisition of genes in the form of pathogenicity islands distinguishes pathogenic isolates from nonpathogens. We therefore sequenced a highly virulent strain of P. aeruginosa, PA14, and compared it with a previously sequenced (and less pathogenic) strain, PAO1, to identify novel virulence genes.
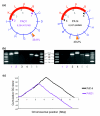
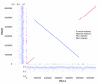
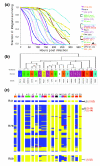
Results: The PA14 and PAO1 genomes are remarkably similar, although PA14 has a slightly larger genome (6.5 megabses [Mb]) than does PAO1 (6.3 Mb). We identified 58 PA14 gene clusters that are absent in PAO1 to determine which of these genes, if any, contribute to its enhanced virulence in a Caenorhabditis elegans pathogenicity model. First, we tested 18 additional diverse strains in the C. elegans model and observed a wide range of pathogenic potential; however, genotyping these strains using a custom microarray showed that the presence of PA14 genes that are absent in PAO1 did not correlate with the virulence of these strains. Second, we utilized a full-genome nonredundant mutant library of PA14 to identify five genes (absent in PAO1) required for C. elegans killing. Surprisingly, although these five genes are present in many other P. aeruginosa strains, they do not correlate with virulence in C. elegans.
Conclusion: Genes required for pathogenicity in one strain of P. aeruginosa are neither required for nor predictive of virulence in other strains. We therefore propose that virulence in this organism is both multifactorial and combinatorial, the result of a pool of pathogenicity-related genes that interact in various combinations in different genetic backgrounds.
Figures

References
-
- Doring D. Pseudomonas aeruginosa as an opportunistic pathogen. New York: Plenum Press; 1993. Chronic Pseudomonas aeruginosa lung infection in cystic fibrosis patients. pp. 245–273.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

