MIMOX: a web tool for phage display based epitope mapping
- PMID: 17038191
- PMCID: PMC1618411
- DOI: 10.1186/1471-2105-7-451
MIMOX: a web tool for phage display based epitope mapping
Abstract
Background: Phage display is widely used in basic research such as the exploration of protein-protein interaction sites and networks, and applied research such as the development of new drugs, vaccines, and diagnostics. It has also become a promising method for epitope mapping. Research on new algorithms that assist and automate phage display based epitope mapping has attracted many groups. Most of the existing tools have not been implemented as an online service until now however, making it less convenient for the community to access, utilize, and evaluate them.
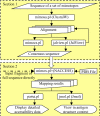
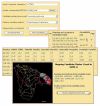
Results: We present MIMOX, a free web tool that helps to map the native epitope of an antibody based on one or more user supplied mimotopes and the antigen structure. MIMOX was coded in Perl using modules from the Bioperl project. It has two sections. In the first section, MIMOX provides a simple interface for ClustalW to align a set of mimotopes. It also provides a simple statistical method to derive the consensus sequence and embeds JalView as a Java applet to view and manage the alignment. In the second section, MIMOX can map a single mimotope or a consensus sequence of a set of mimotopes, on to the corresponding antigen structure and search for all of the clusters of residues that could represent the native epitope. NACCESS is used to evaluate the surface accessibility of the candidate clusters; and Jmol is embedded to view them interactively in their 3D context. Initial case studies show that MIMOX can reproduce mappings from existing tools such as FINDMAP and 3DEX, as well as providing novel, rational results.
Conclusion: A web-based tool called MIMOX has been developed for phage display based epitope mapping. As a publicly available online service in this area, it is convenient for the community to access, utilize, and evaluate, complementing other existing programs. MIMOX is freely available at http://web.kuicr.kyoto-u.ac.jp/~hjian/mimox.
Figures


References
-
- Tong AH, Drees B, Nardelli G, Bader GD, Brannetti B, Castagnoli L, Evangelista M, Ferracuti S, Nelson B, Paoluzi S, et al. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science. 2002;295:321–324. doi: 10.1126/science.1064987. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

