Structural basis for mRNA and tRNA positioning on the ribosome
- PMID: 17038497
- PMCID: PMC1635088
- DOI: 10.1073/pnas.0607541103
Structural basis for mRNA and tRNA positioning on the ribosome
Erratum in
- Proc Natl Acad Sci U S A. 2006 Dec 26;103(52):19931. Cate, Jamie H Doudna [corrected to Cate, Jamie H D]
Abstract
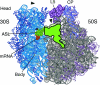
Protein synthesis requires the accurate positioning of mRNA and tRNA in the peptidyl-tRNA site of the ribosome. Here we describe x-ray crystal structures of the intact bacterial ribosome from Escherichia coli in a complex with mRNA and the anticodon stem-loop of P-site tRNA. At 3.5-A resolution, these structures reveal rearrangements in the intact ribosome that clamp P-site tRNA and mRNA on the small ribosomal subunit. Binding of the anticodon stem-loop of P-site tRNA to the ribosome is sufficient to lock the head of the small ribosomal subunit in a single conformation, thereby preventing movement of mRNA and tRNA before mRNA decoding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

