A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses
- PMID: 17039602
- PMCID: PMC1762135
- DOI: 10.1016/j.virol.2006.04.043
A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses
Abstract
HIV-1 subtype C is the most common HIV-1 group M subtype in Africa and many parts of Asia. However, to date HIV-1 vaccine candidate immunogens have not induced potent and broadly neutralizing antibodies against subtype C primary isolates. We have used a centralized gene strategy to address HIV-1 diversity and generated a group M consensus envelope gene with shortened consensus variable loops (CON-S) for comparative studies with wild-type (WT) Env immunogens. Our results indicate that the consensus HIV-1 group M CON-S Env elicited cross-subtype neutralizing antibodies of similar or greater breadth and titer than the WT Envs tested, indicating the utility of a centralized gene strategy. Our study also shows the feasibility of iterative improvements in Env immunogenicity by rational design of centralized genes.
Figures
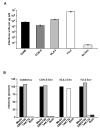








References
-
- Alam S, Paleos C, Liao HX, Scearce R, Robinson J, Haynes B. An inducible HIV type 1 gp41 HR-2 peptide-binding site on HIV type 1 envelope gp120. AIDS Res Hum Retroviruses. 2004;20(8):836–845. - PubMed
-
- Barnett S, Lu S, Srivastava I, Cherpelis S, Gettie A, Blanchard J, Wang S, Mboudjeka I, Leung L, Lian Y, Fong A, Buckner C, Ly A, Hilt S, Ulmer J, Wild C, Mascola J, Stamatatos L. The Ability of an Oligomeric Human Immunodeficiency Virus Type 1 (HIV-1) Envelope Antigen To Elicit Neutralizing Antibodies against Primary HIV-1 Isolates Is Improved following Partial Deletion of the Second Hypervariable Region. J Virol. 2001;75:5526–5540. - PMC - PubMed
-
- Bures R, Gaitan A, Zhu T, Graziosi C, McGrath KM, Tartaglia J, Caudrelier P, El Habib R, Klein M, Lazzarin A, Stablein DM, Deers M, Corey L, Greenberg ML, Schwartz DH, Montefiori DC. Immunization with recombinant canarypox vectors expressing membrane-anchored gp120 followed by gp160 protein boosting fails to generate antibodies that neutralize R5 primary isolates of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2000;16:2019–2035. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21AI55386/AI/NIAID NIH HHS/United States
- P01 AI061734/AI/NIAID NIH HHS/United States
- N01 AI085338/AI/NIAID NIH HHS/United States
- P01 AI52816/AI/NIAID NIH HHS/United States
- P30 AI51445/AI/NIAID NIH HHS/United States
- N01 AI 85338/AI/NIAID NIH HHS/United States
- P0-1, AI061734/AI/NIAID NIH HHS/United States
- P30 AI051445/AI/NIAID NIH HHS/United States
- P01 AI052816/AI/NIAID NIH HHS/United States
- P01 AI 28147/AI/NIAID NIH HHS/United States
- P01 AI046023/AI/NIAID NIH HHS/United States
- P01 AI028147/AI/NIAID NIH HHS/United States
- R21 AI055386/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
