Characterization of a nucleocapsid-like region and of two distinct primer tRNALys,2 binding sites in the endogenous retrovirus Gypsy
- PMID: 17040893
- PMCID: PMC1635307
- DOI: 10.1093/nar/gkl722
Characterization of a nucleocapsid-like region and of two distinct primer tRNALys,2 binding sites in the endogenous retrovirus Gypsy
Abstract
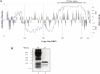
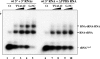
Mobile LTR-retroelements comprising retroviruses and LTR-retrotransposons form a large part of eukaryotic genomes. Their mode of replication and abundance favour the notion that they are major actors in eukaryote evolution. The Gypsy retroelement can spread in the germ line of the fruit fly Drosophila melanogaster via both env-independent and env-dependent processes. Thus, Gypsy is both an active retrotransposon and an infectious retrovirus resembling the gammaretrovirus MuLV. However, unlike gammaretroviruses, the Gypsy Gag structural precursor is not processed into Matrix, Capsid and Nucleocapsid (NC) proteins. In contrast, it has features in common with Gag of the ancient yeast TY1 retroelement. These characteristics of Gypsy make it a very interesting model to study replication of a retroelement at the frontier between ancient retrotransposons and retroviruses. We investigated Gypsy replication using an in vitro model system and transfection of insect cells. Results show that an unstructured domain of Gypsy Gag has all the properties of a retroviral NC. This NC-like peptide forms ribonucleoparticle-like complexes upon binding Gypsy RNA and directs the annealing of primer tRNA(Lys,2) to two distinct primer binding sites (PBS) at the genome 5' and 3' ends. Only the 5' PBS is indispensable for cDNA synthesis in vitro and in Drosophila cells.
Figures






References
-
- Boeke J.D., Stoye J.P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin J.M., Hughes S.H., Varmus H.E., editors. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 343–435. - PubMed
-
- Kazazian H.H., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. - PubMed
-
- Darlix J.L., Lapadat-Tapolsky M., de Rocquigny H., Roques B.P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 1995;254:523–537. - PubMed
-
- Darlix J.L., Cristofari G., Rau M., Pechoux C., Berthoux L., Roques B. Nucleocapsid protein of human immunodeficiency virus as a model protein with chaperoning functions and as a target for antiviral drugs. Adv. Pharmacol. 2000;48:345–372. - PubMed
-
- Rein A., Henderson L.E., Levin J.G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

