Beta-Catenin stabilizes cyclooxygenase-2 mRNA by interacting with AU-rich elements of 3'-UTR
- PMID: 17040897
- PMCID: PMC1636482
- DOI: 10.1093/nar/gkl698
Beta-Catenin stabilizes cyclooxygenase-2 mRNA by interacting with AU-rich elements of 3'-UTR
Abstract
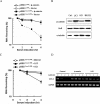
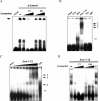
Cyclooxygenase-2 (COX-2) mRNA is induced in the majority of human colorectal carcinomas. Transcriptional regulation plays a key role in COX-2 expression in human colon carcinoma cells, but post-transcriptional regulation of its mRNA is also critical for tumorigenesis. Expression of COX-2 mRNA is regulated by various cytokines, growth factors and other signals. beta-Catenin, a key transcription factor in the Wnt signal pathway, activates transcription of COX-2. Here we found that COX-2 mRNA was also substantially stabilized by activating beta-catenin in NIH3T3 and 293T cells. We identified the beta-catenin-responsive element in the proximal region of the COX-2 3'-untranslated region (3'-UTR) and showed that beta-catenin interacted with AU-rich elements (ARE) of 3'-UTR in vitro and in vivo. Interestingly, beta-catenin induced the cytoplasmic localization of the RNA stabilizing factor, HuR, which may bind to beta-catenin in an RNA-mediated complex and facilitate beta-catenin-dependent stabilization of COX-2 mRNA. Taken together, we provided evidences for beta-catenin as an RNA-binding factor and a regulator of stabilization of COX-2 mRNA.
Figures




Similar articles
-
Functional antagonism between RNA binding proteins HuR and CUGBP2 determines the fate of COX-2 mRNA translation.Gastroenterology. 2007 Mar;132(3):1055-65. doi: 10.1053/j.gastro.2006.12.031. Epub 2006 Dec 19. Gastroenterology. 2007. PMID: 17383427
-
HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3' untranslated regions of cytokine and angiogenic factor mRNAs.Cancer Res. 2001 Mar 1;61(5):2154-61. Cancer Res. 2001. PMID: 11280780
-
AU-rich elements and alternative splicing in the beta-catenin 3'UTR can influence the human beta-catenin mRNA stability.Exp Cell Res. 2006 Jul 15;312(12):2367-78. doi: 10.1016/j.yexcr.2006.03.029. Epub 2006 Apr 15. Exp Cell Res. 2006. PMID: 16696969
-
Complexity of COX-2 gene regulation.Biochem Soc Trans. 2008 Jun;36(Pt 3):543-5. doi: 10.1042/BST0360543. Biochem Soc Trans. 2008. PMID: 18482003 Review.
-
The role of transforming growth factor beta signaling in messenger RNA stability.Growth Factors. 2006 Mar;24(1):1-11. doi: 10.1080/08977190500365995. Growth Factors. 2006. PMID: 16393690 Review.
Cited by
-
Crosstalk between Wnt Signaling and RNA Processing in Colorectal Cancer.J Cancer. 2013;4(2):96-103. doi: 10.7150/jca.5470. Epub 2013 Jan 5. J Cancer. 2013. PMID: 23386908 Free PMC article.
-
Beta-catenin/HuR post-transcriptional machinery governs cancer stem cell features in response to hypoxia.PLoS One. 2013 Nov 15;8(11):e80742. doi: 10.1371/journal.pone.0080742. eCollection 2013. PLoS One. 2013. PMID: 24260469 Free PMC article.
-
RNA localization and polarity: from A(PC) to Z(BP).Trends Cell Biol. 2009 Apr;19(4):156-64. doi: 10.1016/j.tcb.2009.02.001. Epub 2009 Feb 27. Trends Cell Biol. 2009. PMID: 19251418 Free PMC article. Review.
-
Loss of 15-hydroxyprostaglandin dehydrogenase increases prostaglandin E2 in pancreatic tumors.Pancreas. 2010 Apr;39(3):332-9. doi: 10.1097/MPA.0b013e3181baecbe. Pancreas. 2010. PMID: 19820419 Free PMC article.
-
A subpopulation of smooth muscle cells, derived from melanocyte-competent precursors, prevents patent ductus arteriosus.PLoS One. 2013;8(1):e53183. doi: 10.1371/journal.pone.0053183. Epub 2013 Jan 31. PLoS One. 2013. PMID: 23382837 Free PMC article.
References
-
- Maniatis T., Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. - PubMed
-
- Orphanides G., Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. - PubMed
-
- Briata P., Ilengo C., Corte G., Moroni C., Rosenfeld M.G., Chen C.Y., Gherzi R. The Wnt/β-catenin→Pitx2 pathway controls the turnover of Pitx2 and other unstable mRNAs. Mol. Cell. 2003;12:1201–1211. - PubMed
-
- Lerga A., Hallier M., Delva L., Orvain C., Gallais I., Marie J., Moreau-Gachelin F. Identification of an RNA binding specificity for the potential splicing factor TLS. J. Biol. Chem. 2001;276:6807–6816. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

