Common plantain. A collection of expressed sequence tags from vascular tissue and a simple and efficient transformation method
- PMID: 17041024
- PMCID: PMC1676067
- DOI: 10.1104/pp.106.089169
Common plantain. A collection of expressed sequence tags from vascular tissue and a simple and efficient transformation method
Abstract
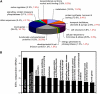
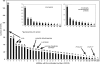
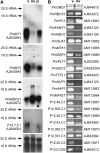
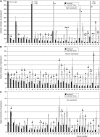
The vascular tissue of higher plants consists of specialized cells that differ from all other cells with respect to their shape and size, their organellar composition, their extracellular matrix, the type of their plasmodesmata, and their physiological functions. Intact and pure vascular tissue can be isolated easily and rapidly from leaf blades of common plantain (Plantago major), a plant that has been used repeatedly for molecular studies of phloem transport. Here, we present a transcriptome analysis based on 5,900 expressed sequence tags (ESTs) and 3,247 independent mRNAs from the Plantago vasculature. The vascular specificity of these ESTs was confirmed by the identification of well-known phloem or xylem marker genes. Moreover, reverse transcription-polymerase chain reaction, macroarray, and northern analyses revealed genes and metabolic pathways that had previously not been described to be vascular specific. Moreover, common plantain transformation was established and used to confirm the vascular specificity of a Plantago promoter-beta-glucuronidase construct in transgenic Plantago plants. Eventually, the applicability and usefulness of the obtained data were also demonstrated for other plant species. Reporter gene constructs generated with promoters from Arabidopsis (Arabidopsis thaliana) homologs of newly identified Plantago vascular ESTs revealed vascular specificity of these genes in Arabidopsis as well. The presented vascular ESTs and the newly developed transformation system represent an important tool for future studies of functional genomics in the common plantain vasculature.
Figures


References
-
- Aitchitt M, Ainsworth CC, Thangavelu M (1993) A rapid and efficient method for the extraction of total DNA from mature leaves of the date palm (Phoenix dactylifera L.). Plant Mol Biol Rep 11: 317–319
-
- Asano T, Masumura T, Kusano H, Kikuchi S, Kurita A, Shimada H, Kadowaki K (2002) Construction of a specialized cDNA library from plant cells isolated by laser capture microdissection: toward comprehensive analysis of the genes expressed in the rice phloem. Plant J 32: 401–408 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

