Analysis of proline reduction in the nosocomial pathogen Clostridium difficile
- PMID: 17041035
- PMCID: PMC1698225
- DOI: 10.1128/JB.01370-06
Analysis of proline reduction in the nosocomial pathogen Clostridium difficile
Abstract
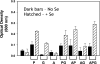
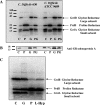
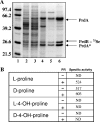
Clostridium difficile, a proteolytic strict anaerobe, has emerged as a clinically significant nosocomial pathogen in recent years. Pathogenesis is due to the production of lethal toxins, A and B, members of the large clostridial cytotoxin family. Although it has been established that alterations in the amino acid content of the growth medium affect toxin production, the molecular mechanism for this observed effect is not yet known. Since there is a paucity of information on the amino acid fermentation pathways used by this pathogen, we investigated whether Stickland reactions might be at the heart of its bioenergetic pathways. Growth of C. difficile on Stickland pairs yielded large increases in cell density in a limiting basal medium, demonstrating that these reactions are tied to ATP production. Selenium supplementation was required for this increase in cell yield. Analysis of genome sequence data reveals genes encoding the protein components of two key selenoenzyme reductases, glycine reductase and d-proline reductase (PR). These selenoenzymes were expressed upon the addition of the corresponding Stickland acceptor (glycine, proline, or hydroxyproline). Purification of the selenoenzyme d-proline reductase revealed a mixed complex of PrdA and PrdB (SeCys-containing) proteins. PR utilized only d-proline but not l-hydroxyproline, even in the presence of an expressed and purified proline racemase. PR was found to be independent of divalent cations, and zinc was a potent inhibitor of PR. These results show that Stickland reactions are key to the growth of C. difficile and that the mechanism of PR may differ significantly from that of previously studied PR from nonpathogenic species.
Figures

Similar articles
-
Proline-dependent regulation of Clostridium difficile Stickland metabolism.J Bacteriol. 2013 Feb;195(4):844-54. doi: 10.1128/JB.01492-12. Epub 2012 Dec 7. J Bacteriol. 2013. PMID: 23222730 Free PMC article.
-
d-Proline Reductase Underlies Proline-Dependent Growth of Clostridioides difficile.J Bacteriol. 2022 Aug 16;204(8):e0022922. doi: 10.1128/jb.00229-22. Epub 2022 Jul 13. J Bacteriol. 2022. PMID: 35862761 Free PMC article.
-
Role of the global regulator Rex in control of NAD+ -regeneration in Clostridioides (Clostridium) difficile.Mol Microbiol. 2019 Jun;111(6):1671-1688. doi: 10.1111/mmi.14245. Epub 2019 Apr 2. Mol Microbiol. 2019. PMID: 30882947 Free PMC article.
-
Glycine metabolism in anaerobes.Antonie Van Leeuwenhoek. 1994;66(1-3):223-37. doi: 10.1007/BF00871641. Antonie Van Leeuwenhoek. 1994. PMID: 7747933 Review.
-
Integration of metabolism and virulence in Clostridium difficile.Res Microbiol. 2015 May;166(4):375-83. doi: 10.1016/j.resmic.2014.10.002. Epub 2014 Oct 15. Res Microbiol. 2015. PMID: 25445566 Free PMC article. Review.
Cited by
-
Clostridium sporogenes uses reductive Stickland metabolism in the gut to generate ATP and produce circulating metabolites.Nat Microbiol. 2022 May;7(5):695-706. doi: 10.1038/s41564-022-01109-9. Epub 2022 May 2. Nat Microbiol. 2022. PMID: 35505245 Free PMC article.
-
Reconsidering the in vivo functions of Clostridial Stickland amino acid fermentations.Anaerobe. 2022 Aug;76:102600. doi: 10.1016/j.anaerobe.2022.102600. Epub 2022 Jun 13. Anaerobe. 2022. PMID: 35709938 Free PMC article. Review.
-
Metabolism the Difficile Way: The Key to the Success of the Pathogen Clostridioides difficile.Front Microbiol. 2019 Feb 15;10:219. doi: 10.3389/fmicb.2019.00219. eCollection 2019. Front Microbiol. 2019. PMID: 30828322 Free PMC article. Review.
-
Iron-sulfur cluster-dependent enzymes and molybdenum-dependent reductases in the anaerobic metabolism of human gut microbes.Metallomics. 2024 Nov 7;16(11):mfae049. doi: 10.1093/mtomcs/mfae049. Metallomics. 2024. PMID: 39504489 Free PMC article. Review.
-
Auranofin: repurposing an old drug for a golden new age.Drugs R D. 2015 Mar;15(1):13-20. doi: 10.1007/s40268-015-0083-y. Drugs R D. 2015. PMID: 25698589 Free PMC article.
References
-
- Andreesen, J. R., M. Wagner, D. Sonntag, M. Kohlstock, C. Harms, T. Gursinsky, J. Jager, T. Parther, U. Kabisch, A. Grantzdorffer, A. Pich, and B. Sohling. 1999. Various functions of selenols and thiols in anaerobic gram-positive, amino acids-utilizing bacteria. Biofactors 10:263-270. - PubMed
-
- Barker, H. A. 1961. Fermentation of nitrogenous organic compounds, p. 151-188. In I. C. Gunsalus and R. Y. Stanier (ed.), The bacteria, vol. 2. Academic Press, New York, N.Y.
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Britz, M. L., and R. G. Wilkinson. 1982. Leucine dissimilation to isovaleric and isocaproic acids by cell suspensions of amino acid fermenting anaerobes: the Stickland reaction revisited. Can. J. Microbiol. 28:291-300. - PubMed
-
- Cases, J., V. Vacchina, A. Napolitano, B. Caporiccio, P. Besancon, R. Lobinski, and J. M. Rouanet. 2001. Selenium from selenium-rich Spirulina is less bioavailable than selenium from sodium selenite and selenomethionine in selenium-deficient rats. J. Nutr. 131:2343-2350. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

