Agrobacterium tumefaciens soxR is involved in superoxide stress protection and also directly regulates superoxide-inducible expression of itself and a target gene
- PMID: 17041041
- PMCID: PMC1698218
- DOI: 10.1128/JB.00856-06
Agrobacterium tumefaciens soxR is involved in superoxide stress protection and also directly regulates superoxide-inducible expression of itself and a target gene
Abstract
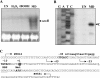
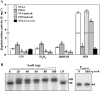
Inactivation of Agrobacterium tumefaciens soxR increases sensitivity to superoxide generators. soxR expression is highly induced by superoxide stress and is autoregulated. SoxR also directly regulates the superoxide-inducible expression of atu5152. Taken together, the physiological role of soxR and the mechanism by which it regulates expression of target genes make the A. tumefaciens SoxR system different from other bacterial systems.
Figures
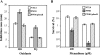

Similar articles
-
The transcription factors ActR and SoxR differentially affect the phenazine tolerance of Agrobacterium tumefaciens.Mol Microbiol. 2019 Jul;112(1):199-218. doi: 10.1111/mmi.14263. Epub 2019 May 3. Mol Microbiol. 2019. PMID: 31001852 Free PMC article.
-
Oxidant-inducible resistance to hydrogen peroxide killing in Agrobacterium tumefaciens requires the global peroxide sensor-regulator OxyR and KatA.FEMS Microbiol Lett. 2003 Aug 8;225(1):167-72. doi: 10.1016/S0378-1097(03)00511-1. FEMS Microbiol Lett. 2003. PMID: 12900037
-
DnaK chaperone-mediated control of activity of a sigma(32) homolog (RpoH) plays a major role in the heat shock response of Agrobacterium tumefaciens.J Bacteriol. 2001 Sep;183(18):5302-10. doi: 10.1128/JB.183.18.5302-5310.2001. J Bacteriol. 2001. PMID: 11514513 Free PMC article.
-
Lysine decarboxylase expression by Vibrio vulnificus is induced by SoxR in response to superoxide stress.J Bacteriol. 2006 Dec;188(24):8586-92. doi: 10.1128/JB.01084-06. Epub 2006 Sep 29. J Bacteriol. 2006. PMID: 17012399 Free PMC article.
-
Sensing and protecting against superoxide stress in Escherichia coli--how many ways are there to trigger soxRS response?Redox Rep. 2000;5(5):287-93. doi: 10.1179/135100000101535825. Redox Rep. 2000. PMID: 11145103 Review.
Cited by
-
The transcription factors ActR and SoxR differentially affect the phenazine tolerance of Agrobacterium tumefaciens.Mol Microbiol. 2019 Jul;112(1):199-218. doi: 10.1111/mmi.14263. Epub 2019 May 3. Mol Microbiol. 2019. PMID: 31001852 Free PMC article.
-
Physiological and expression analyses of Agrobacterium tumefaciens trxA, encoding thioredoxin.J Bacteriol. 2007 Sep;189(17):6477-81. doi: 10.1128/JB.00623-07. Epub 2007 Jun 15. J Bacteriol. 2007. PMID: 17573482 Free PMC article.
-
The catalase-peroxidase KatG is required for virulence of Xanthomonas campestris pv. campestris in a host plant by providing protection against low levels of H2O2.J Bacteriol. 2009 Dec;191(23):7372-7. doi: 10.1128/JB.00788-09. Epub 2009 Sep 25. J Bacteriol. 2009. PMID: 19783631 Free PMC article.
-
Cellular defenses against superoxide and hydrogen peroxide.Annu Rev Biochem. 2008;77:755-76. doi: 10.1146/annurev.biochem.77.061606.161055. Annu Rev Biochem. 2008. PMID: 18173371 Free PMC article. Review.
-
Redox-active antibiotics control gene expression and community behavior in divergent bacteria.Science. 2008 Aug 29;321(5893):1203-6. doi: 10.1126/science.1160619. Science. 2008. PMID: 18755976 Free PMC article.
References
-
- Claros, M. G., and G. von Heijne. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10:685-686. - PubMed
-
- Demple, B. 1996. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon—a review. Gene 179:53-57. - PubMed
-
- Demple, B., E. Hidalgo, and H. Ding. 1999. Transcriptional regulation via redox-sensitive iron-sulphur centres in an oxidative stress response. Biochem. Soc. Symp. 64:119-128. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

