Expression of the pyr operon of Lactobacillus plantarum is regulated by inorganic carbon availability through a second regulator, PyrR2, homologous to the pyrimidine-dependent regulator PyrR1
- PMID: 17041052
- PMCID: PMC1698236
- DOI: 10.1128/JB.00985-06
Expression of the pyr operon of Lactobacillus plantarum is regulated by inorganic carbon availability through a second regulator, PyrR2, homologous to the pyrimidine-dependent regulator PyrR1
Abstract
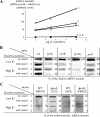
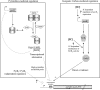
Inorganic carbon (IC), such as bicarbonate or carbon dioxide, stimulates the growth of Lactobacillus plantarum. At low IC levels, one-third of natural isolated L. plantarum strains are nutritionally dependent on exogenous arginine and pyrimidine, a phenotype previously defined as high-CO2-requiring (HCR) prototrophy. IC enrichment significantly decreased the amounts of the enzymes in the pyrimidine biosynthetic pathway encoded by the pyrR1BCAa1Ab1DFE operon, as demonstrated by proteomic analysis. Northern blot and reverse transcription-PCR experiments demonstrated that IC levels regulated pyr genes mainly at the level of transcription or RNA stability. Two putative PyrR regulators with 62% amino acid identity are present in the L. plantarum genome. PyrR1 is an RNA-binding protein that regulates the pyr genes in response to pyrimidine availability by a mechanism of transcriptional attenuation. In this work, the role of PyrR2 was investigated by allelic gene replacement. Unlike the pyrR1 mutant, the DeltapyrR2 strain acquired a demand for both pyrimidines and arginine unless bicarbonate or CO2 was present at high concentrations, which is known as an HCR phenotype. Analysis of the IC- and pyrimidine-mediated regulation in pyrR1 and pyrR2 mutants suggested that only PyrR2 positively regulates the expression levels of the pyr genes in response to IC levels but had no effect on pyrimidine-mediated repression. A model is proposed for the respective roles of PyrR1 and PyrR2 in the pyr regulon expression.
Figures



References
-
- Arsène-Ploetze, F., and F. Bringel. 2004. Role of inorganic carbon on lactic acid bacteria metabolism. Lait 84:45-59.
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 72:248-254. - PubMed
-
- Bringel, F., and J. C. Hubert. 2003. Extent of genetic lesions of the arginine and pyrimidine biosynthetic pathways in Lactobacillus plantarum, L. paraplantarum, L. pentosus, and L. casei: prevalence of CO2 dependent auxotrophs and characterization of deficient arg genes in L. plantarum. Appl. Environ. Microbiol. 69:2674-2683. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

