Malonyl-coenzyme A reductase in the modified 3-hydroxypropionate cycle for autotrophic carbon fixation in archaeal Metallosphaera and Sulfolobus spp
- PMID: 17041055
- PMCID: PMC1698253
- DOI: 10.1128/JB.00987-06
Malonyl-coenzyme A reductase in the modified 3-hydroxypropionate cycle for autotrophic carbon fixation in archaeal Metallosphaera and Sulfolobus spp
Abstract
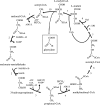
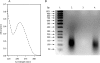
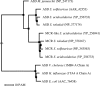
Autotrophic members of the Sulfolobales (Crenarchaeota) contain acetyl-coenzyme A (CoA)/propionyl-CoA carboxylase as the CO2 fixation enzyme and use a modified 3-hydroxypropionate cycle to assimilate CO2 into cell material. In this central metabolic pathway malonyl-CoA, the product of acetyl-CoA carboxylation, is further reduced to 3-hydroxypropionate. Extracts of Metallosphaera sedula contained NADPH-specific malonyl-CoA reductase activity that was 10-fold up-regulated under autotrophic growth conditions. Malonyl-CoA reductase was partially purified and studied. Based on N-terminal amino acid sequencing the corresponding gene was identified in the genome of the closely related crenarchaeum Sulfolobus tokodaii. The Sulfolobus gene was cloned and heterologously expressed in Escherichia coli, and the recombinant protein was purified and studied. The enzyme catalyzes the following reaction: malonyl-CoA + NADPH + H+ --> malonate-semialdehyde + CoA + NADP+. In its native state it is associated with small RNA. Its activity was stimulated by Mg2+ and thiols and inactivated by thiol-blocking agents, suggesting the existence of a cysteine adduct in the course of the catalytic cycle. The enzyme was specific for NADPH (Km = 25 microM) and malonyl-CoA (Km = 40 microM). Malonyl-CoA reductase has 38% amino acid sequence identity to aspartate-semialdehyde dehydrogenase, suggesting a common ancestor for both proteins. It does not exhibit any significant similarity with malonyl-CoA reductase from Chloroflexus aurantiacus. This shows that the autotrophic pathway in Chloroflexus and Sulfolobaceae has evolved convergently and that these taxonomic groups have recruited different genes to bring about similar metabolic processes.
Figures




Similar articles
-
3-Hydroxypropionyl-coenzyme A synthetase from Metallosphaera sedula, an enzyme involved in autotrophic CO2 fixation.J Bacteriol. 2008 Feb;190(4):1383-9. doi: 10.1128/JB.01593-07. Epub 2007 Dec 28. J Bacteriol. 2008. PMID: 18165310 Free PMC article.
-
Presence of acetyl coenzyme A (CoA) carboxylase and propionyl-CoA carboxylase in autotrophic Crenarchaeota and indication for operation of a 3-hydroxypropionate cycle in autotrophic carbon fixation.J Bacteriol. 1999 Feb;181(4):1088-98. doi: 10.1128/JB.181.4.1088-1098.1999. J Bacteriol. 1999. PMID: 9973333 Free PMC article.
-
Malonyl-coenzyme A reductase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO(2) fixation.J Bacteriol. 2002 May;184(9):2404-10. doi: 10.1128/JB.184.9.2404-2410.2002. J Bacteriol. 2002. PMID: 11948153 Free PMC article.
-
Occurrence, biochemistry and possible biotechnological application of the 3-hydroxypropionate cycle.Appl Microbiol Biotechnol. 2004 Jun;64(5):605-10. doi: 10.1007/s00253-003-1540-z. Epub 2004 Feb 28. Appl Microbiol Biotechnol. 2004. PMID: 14997352 Review.
-
Unfamiliar metabolic links in the central carbon metabolism.J Biotechnol. 2014 Dec 20;192 Pt B:314-22. doi: 10.1016/j.jbiotec.2014.02.015. Epub 2014 Feb 24. J Biotechnol. 2014. PMID: 24576434 Review.
Cited by
-
Exploiting microbial hyperthermophilicity to produce an industrial chemical, using hydrogen and carbon dioxide.Proc Natl Acad Sci U S A. 2013 Apr 9;110(15):5840-5. doi: 10.1073/pnas.1222607110. Epub 2013 Mar 25. Proc Natl Acad Sci U S A. 2013. PMID: 23530213 Free PMC article.
-
Comparative Genomic Analysis Reveals the Metabolism and Evolution of the Thermophilic Archaeal Genus Metallosphaera.Front Microbiol. 2020 Jun 19;11:1192. doi: 10.3389/fmicb.2020.01192. eCollection 2020. Front Microbiol. 2020. PMID: 32655516 Free PMC article.
-
Role of 4-hydroxybutyrate-CoA synthetase in the CO2 fixation cycle in thermoacidophilic archaea.J Biol Chem. 2013 Feb 8;288(6):4012-22. doi: 10.1074/jbc.M112.413195. Epub 2012 Dec 20. J Biol Chem. 2013. PMID: 23258541 Free PMC article.
-
Microbial CO2 fixation and biotechnology in reducing industrial CO2 emissions.Arch Microbiol. 2022 Jan 21;204(2):149. doi: 10.1007/s00203-021-02677-w. Arch Microbiol. 2022. PMID: 35061105 Review.
-
The divergence and natural selection of autocatalytic primordial metabolic systems.Orig Life Evol Biosph. 2013 Jun;43(3):263-81. doi: 10.1007/s11084-013-9340-7. Epub 2013 Jul 17. Orig Life Evol Biosph. 2013. PMID: 23860777
References
-
- Aiba, H., S. Adhya, and B. de Crombrugghe. 1981. Evidence for two functional gal promotors in intact Escherichia coli. J. Biol. Chem. 256:11905-11910. - PubMed
-
- Alber, B. E., and G. Fuchs. 2002. Propionyl-coenzyme A synthase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation. J. Biol. Chem. 277:12137-12143. - PubMed
-
- Biellmann, J. F., P. Eid, and C. Hirth. 1980. Affinity labeling of the Escherichia coli aspartate-beta-semialdehyde dehydrogenase with an alkylating coenzyme analogue. Half-site reactivity and competition with the substrate alkylating analogue. Eur. J. Biochem. 104:65-69. - PubMed
-
- Blanco, J., R. A. Moore, C. R. Faehnle, and R. E. Viola. 2004. Critical catalytic functional groups in the mechanism of aspartate-β-semialdehyde dehydrogenase. Acta Crystallogr. Sect. D Biol. Crystallogr. 60:1808-1815. - PubMed
-
- Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

