Identification of a gene negatively affecting antibiotic production and morphological differentiation in Streptomyces coelicolor A3(2)
- PMID: 17041057
- PMCID: PMC1698255
- DOI: 10.1128/JB.00933-06
Identification of a gene negatively affecting antibiotic production and morphological differentiation in Streptomyces coelicolor A3(2)
Abstract
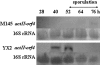
SC7A1 is a cosmid with an insert of chromosomal DNA from Streptomyces coelicolor A3(2). Its insertion into the chromosome of S. coelicolor strains caused a duplication of a segment of ca. 40 kb and delayed actinorhodin antibiotic production and sporulation, implying that SC7A1 carried a gene negatively affecting these processes. The subcloning of SC7A1 insert DNA resulted in the identification of the open reading frame SCO5582 as nsdA, a gene negatively affecting Streptomyces differentiation. The disruption of chromosomal nsdA caused the overproduction of spores and of three of four known S. coelicolor antibiotics of quite different chemical types. In at least one case (that of actinorhodin), this was correlated with premature expression of a pathway-specific regulatory gene (actII-orf4), implying that nsdA in the wild-type strain indirectly repressed the expression of the actinorhodin biosynthesis cluster. nsdA expression was up-regulated upon aerial mycelium initiation and was strongest in the aerial mycelium. NsdA has DUF921, a Streptomyces protein domain of unknown function and a conserved SXR site. A site-directed mutation (S458A) in this site in NsdA abolished its function. Blast searching showed that NsdA homologues are present in some Streptomyces genomes. Outside of streptomycetes, NsdA-like proteins have been found in several actinomycetes. The disruption of the nsdA-like gene SCO4114 had no obvious phenotypic effects on S. coelicolor. The nsdA orthologue SAV2652 in S. avermitilis could complement the S. coelicolor nsdA-null mutant phenotype.
Figures






Similar articles
-
A Regulatory Gene SCO2140 is Involved in Antibiotic Production and Morphological Differentiation of Streptomyces coelicolor A3(2).Curr Microbiol. 2016 Aug;73(2):196-201. doi: 10.1007/s00284-016-1050-8. Epub 2016 Apr 25. Curr Microbiol. 2016. PMID: 27113590
-
NsdB, a TPR-like-domain-containing protein negatively affecting production of antibiotics in Streptomyces coelicolor A3 (2).Wei Sheng Wu Xue Bao. 2007 Oct;47(5):849-54. Wei Sheng Wu Xue Bao. 2007. PMID: 18062261
-
A putative transglycosylase encoded by SCO4132 influences morphological differentiation and actinorhodin production in Streptomyces coelicolor.Acta Biochim Biophys Sin (Shanghai). 2013 Apr;45(4):296-302. doi: 10.1093/abbs/gmt012. Epub 2013 Feb 11. Acta Biochim Biophys Sin (Shanghai). 2013. PMID: 23403510
-
Production of specialized metabolites by Streptomyces coelicolor A3(2).Adv Appl Microbiol. 2014;89:217-66. doi: 10.1016/B978-0-12-800259-9.00006-8. Adv Appl Microbiol. 2014. PMID: 25131404 Review.
-
The master regulator PhoP coordinates phosphate and nitrogen metabolism, respiration, cell differentiation and antibiotic biosynthesis: comparison in Streptomyces coelicolor and Streptomyces avermitilis.J Antibiot (Tokyo). 2017 May;70(5):534-541. doi: 10.1038/ja.2017.19. Epub 2017 Mar 15. J Antibiot (Tokyo). 2017. PMID: 28293039 Review.
Cited by
-
Genome-Wide Analysis Reveals the Secondary Metabolome in Streptomyces kanasensis ZX01.Genes (Basel). 2017 Nov 30;8(12):346. doi: 10.3390/genes8120346. Genes (Basel). 2017. PMID: 29189712 Free PMC article.
-
Uncovering the prevalence and diversity of integrating conjugative elements in actinobacteria.PLoS One. 2011;6(11):e27846. doi: 10.1371/journal.pone.0027846. Epub 2011 Nov 16. PLoS One. 2011. PMID: 22114709 Free PMC article.
-
LAL regulators SCO0877 and SCO7173 as pleiotropic modulators of phosphate starvation response and actinorhodin biosynthesis in Streptomyces coelicolor.PLoS One. 2012;7(2):e31475. doi: 10.1371/journal.pone.0031475. Epub 2012 Feb 20. PLoS One. 2012. PMID: 22363654 Free PMC article.
-
Identification of a gene from Streptomyces rimosus M527 negatively affecting rimocidin biosynthesis and morphological differentiation.Appl Microbiol Biotechnol. 2020 Dec;104(23):10191-10202. doi: 10.1007/s00253-020-10955-8. Epub 2020 Oct 15. Appl Microbiol Biotechnol. 2020. PMID: 33057790
-
Genome-Wide Mutagenesis Links Multiple Metabolic Pathways with Actinorhodin Production in Streptomyces coelicolor.Appl Environ Microbiol. 2019 Mar 22;85(7):e03005-18. doi: 10.1128/AEM.03005-18. Print 2019 Apr 1. Appl Environ Microbiol. 2019. PMID: 30709825 Free PMC article.
References
-
- Anderson, T. B., P. Brian, and W. C. Champness. 2001. Genetic and transcriptional analysis of absA, an antibiotic gene cluster-linked two-component system that regulates multiple antibiotics in Streptomyces coelicolor. Mol. Microbiol. 39:553-566. - PubMed
-
- Babcock, M. J., and K. E. Kendrick. 1990. Transcriptional and translational features of a sporulation gene of Streptomyces griseus. Gene 95:57-63. - PubMed
-
- Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. - PubMed
-
- Bibb, M. 1996. 1995 Colworth Prize Lecture. The regulation of antibiotic production in Streptomyces coelicolor A3(2). Microbiology 142:1335-1344. - PubMed
-
- Bignell, D. R., J. L. Warawa, J. L. Strap, K. F. Chater, and B. K. Leskiw. 2000. Study of the bldG locus suggests that an anti-anti-sigma factor and an anti-sigma factor may be involved in Streptomyces coelicolor antibiotic production and sporulation. Microbiology 146:2161-2173. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

