Membrane topology mapping of the Na+-pumping NADH: quinone oxidoreductase from Vibrio cholerae by PhoA-green fluorescent protein fusion analysis
- PMID: 17041063
- PMCID: PMC1698230
- DOI: 10.1128/JB.01383-06
Membrane topology mapping of the Na+-pumping NADH: quinone oxidoreductase from Vibrio cholerae by PhoA-green fluorescent protein fusion analysis
Abstract
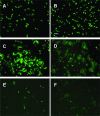
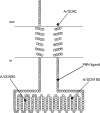
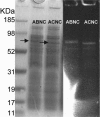
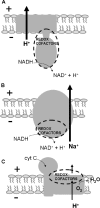
The membrane topologies of the six subunits of Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae were determined by a combination of topology prediction algorithms and the construction of C-terminal fusions. Fusion expression vectors contained either bacterial alkaline phosphatase (phoA) or green fluorescent protein (gfp) genes as reporters of periplasmic and cytoplasmic localization, respectively. A majority of the topology prediction algorithms did not predict any transmembrane helices for NqrA. A lack of PhoA activity when fused to the C terminus of NqrA and the observed fluorescence of the green fluorescent protein C-terminal fusion confirm that this subunit is localized to the cytoplasmic side of the membrane. Analysis of four PhoA fusions for NqrB indicates that this subunit has nine transmembrane helices and that residue T236, the binding site for flavin mononucleotide (FMN), resides in the cytoplasm. Three fusions confirm that the topology of NqrC consists of two transmembrane helices with the FMN binding site at residue T225 on the cytoplasmic side. Fusion analysis of NqrD and NqrE showed almost mirror image topologies, each consisting of six transmembrane helices; the results for NqrD and NqrE are consistent with the topologies of Escherichia coli homologs YdgQ and YdgL, respectively. The NADH, flavin adenine dinucleotide, and Fe-S center binding sites of NqrF were localized to the cytoplasm. The determination of the topologies of the subunits of Na+-NQR provides valuable insights into the location of cofactors and identifies targets for mutagenesis to characterize this enzyme in more detail. The finding that all the redox cofactors are localized to the cytoplasmic side of the membrane is discussed.
Figures




Similar articles
-
Crystallization and preliminary analysis of the NqrA and NqrC subunits of the Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio cholerae.Acta Crystallogr F Struct Biol Commun. 2014 Jul;70(Pt 7):987-92. doi: 10.1107/S2053230X14009881. Epub 2014 Jun 19. Acta Crystallogr F Struct Biol Commun. 2014. PMID: 25005105 Free PMC article.
-
The structure of Na⁺-translocating of NADH:ubiquinone oxidoreductase of Vibrio cholerae: implications on coupling between electron transfer and Na⁺ transport.Biol Chem. 2015 Sep;396(9-10):1015-30. doi: 10.1515/hsz-2015-0128. Biol Chem. 2015. PMID: 26146127 Review.
-
Localization and function of the membrane-bound riboflavin in the Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae.J Biol Chem. 2010 Aug 27;285(35):27088-27099. doi: 10.1074/jbc.M109.071126. Epub 2010 Jun 17. J Biol Chem. 2010. PMID: 20558724 Free PMC article.
-
NqrM (DUF539) Protein Is Required for Maturation of Bacterial Na+-Translocating NADH:Quinone Oxidoreductase.J Bacteriol. 2015 Dec 7;198(4):655-63. doi: 10.1128/JB.00757-15. J Bacteriol. 2015. PMID: 26644436 Free PMC article.
-
The Na+-translocating NADH:quinone oxidoreductase (Na+-NQR): Physiological role, structure and function of a redox-driven, molecular machine.Biochim Biophys Acta Bioenerg. 2024 Nov 1;1865(4):149485. doi: 10.1016/j.bbabio.2024.149485. Epub 2024 Jun 30. Biochim Biophys Acta Bioenerg. 2024. PMID: 38955304 Review.
Cited by
-
Crystallization and preliminary analysis of the NqrA and NqrC subunits of the Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio cholerae.Acta Crystallogr F Struct Biol Commun. 2014 Jul;70(Pt 7):987-92. doi: 10.1107/S2053230X14009881. Epub 2014 Jun 19. Acta Crystallogr F Struct Biol Commun. 2014. PMID: 25005105 Free PMC article.
-
A mutation in Na(+)-NQR uncouples electron flow from Na(+) translocation in the presence of K(+).Biochemistry. 2015 Jan 20;54(2):490-6. doi: 10.1021/bi501266e. Epub 2014 Dec 22. Biochemistry. 2015. PMID: 25486106 Free PMC article.
-
Origin and evolution of the sodium -pumping NADH: ubiquinone oxidoreductase.PLoS One. 2014 May 8;9(5):e96696. doi: 10.1371/journal.pone.0096696. eCollection 2014. PLoS One. 2014. PMID: 24809444 Free PMC article.
-
Membrane topology analysis of HIV-1 envelope glycoprotein gp41.Retrovirology. 2010 Nov 30;7:100. doi: 10.1186/1742-4690-7-100. Retrovirology. 2010. PMID: 21118523 Free PMC article.
-
Requirement of NMB0065 for connecting assembly and export of sialic acid capsular polysaccharides in Neisseria meningitidis.Microbes Infect. 2010 Jun;12(6):476-87. doi: 10.1016/j.micinf.2010.02.009. Epub 2010 Mar 7. Microbes Infect. 2010. PMID: 20215001 Free PMC article.
References
-
- Barquera, B., C. C. Hase, and R. B. Gennis. 2001. Expression and mutagenesis of the NqrC subunit of the NQR respiratory Na(+) pump from Vibrio cholerae with covalently attached FMN. FEBS Lett. 492:45-49. - PubMed
-
- Barquera, B., P. Hellwig, W. Zhou, J. E. Morgan, C. C. Hase, K. K. Gosink, M. Nilges, P. J. Bruesehoff, A. Roth, C. R. Lancaster, and R. B. Gennis. 2002. Purification and characterization of the recombinant Na(+)-translocating NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry 41:3781-3789. - PubMed
-
- Barquera, B., M. J. Nilges, J. E. Morgan, L. Ramirez-Silva, W. Zhou, and R. B. Gennis. 2004. Mutagenesis study of the 2Fe-2S center and the FAD binding site of the Na(+)-translocating NADH:ubiquinone oxidoreductase from Vibrio cholerae. Biochemistry 43:12322-12330. - PubMed
-
- Bertsova, Y., and A. V. Bogachev. 2004. The origin of the sodium-dependent NADH oxidation by the respiratory chain of Klebsiella pneumoniae. FEBS Lett. 563:207-212. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

