Human immunodeficiency virus type 1 Gag polyprotein modulates its own translation
- PMID: 17041220
- PMCID: PMC1641787
- DOI: 10.1128/JVI.02596-05
Human immunodeficiency virus type 1 Gag polyprotein modulates its own translation
Abstract
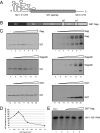
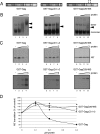
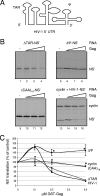
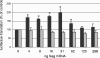
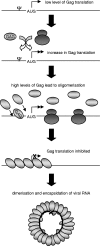
The full-length viral RNA of human immunodeficiency virus type 1 (HIV-1) functions both as the mRNA for the viral structural proteins Gag and Gag/Pol and as the genomic RNA packaged within viral particles. The packaging signal which Gag recognizes to initiate genome encapsidation is in the 5' untranslated region (UTR) of the HIV-1 RNA, which is also the location of translation initiation complex formation. Hence, it is likely that there is competition between the translation and packaging processes. We studied the ability of Gag to regulate translation of its own mRNA. Gag had a bimodal effect on translation from the HIV-1 5' UTR, stimulating translation at low concentrations and inhibiting translation at high concentrations in vitro and in vivo. The inhibition was dependent upon the ability of Gag to bind the packaging signal through its nucleocapsid domain. The stimulatory activity was shown to depend on the matrix domain of Gag. These results suggest that Gag controls the equilibrium between translation and packaging, ensuring production of enough molecules of Gag to make viral particles before encapsidating its genome.
Figures

References
-
- Bedard, K. M., and B. L. Semler. 2004. Regulation of picornavirus gene expression. Microbes Infect. 6:702-713. - PubMed
-
- Borman, A., and R. J. Jackson. 1992. Initiation of translation of human rhinovirus RNA: mapping the internal ribosome entry site. Virology 188:685-696. - PubMed
-
- Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

