At least 50% of human-specific HERV-K (HML-2) long terminal repeats serve in vivo as active promoters for host nonrepetitive DNA transcription
- PMID: 17041225
- PMCID: PMC1641792
- DOI: 10.1128/JVI.00871-06
At least 50% of human-specific HERV-K (HML-2) long terminal repeats serve in vivo as active promoters for host nonrepetitive DNA transcription
Abstract
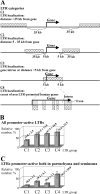
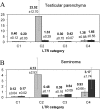
We report the first genome-wide comparison of in vivo promoter activities of a group of human-specific endogenous retroviruses in healthy and cancerous germ line tissues. To this end, we employed a recently developed technique termed genomic repeat expression monitoring. We found that at least 50% of human-specific long terminal repeats (LTRs) possessed promoter activity, and many of them were up- or downregulated in a seminoma. Individual LTRs were expressed at markedly different levels, ranging from approximately 0.001 to approximately 3% of the housekeeping beta-actin gene transcript level. We demonstrated that the main factors affecting the LTR promoter activity were the LTR type (5'-proviral, 3' proviral, or solitary) and position with regard to genes. The averaged promoter strengths of solitary and 3'-proviral LTRs were almost identical in both tissues, whereas 5'-proviral LTRs displayed two- to fivefold higher promoter activities. The relative content of promoter-active LTRs in gene-rich regions was significantly higher than that in gene-poor loci. This content was maximal in those regions where LTRs "overlapped" readthrough transcripts. Although many promoter-active LTRs were mapped near known genes, no clear-cut correlation was observed between transcriptional activities of genes and neighboring LTRs. Our data also suggest a selective suppression of transcription for LTRs located in gene introns.
Figures
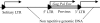



References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinard, K. K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861-868. - PubMed
-
- Buscher, K., U. Trefzer, M. Hofmann, W. Sterry, R. Kurth, and J. Denner. 2005. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 65:4172-4180. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

