Genomics meets HIV-1
- PMID: 17041633
- PMCID: PMC2043131
- DOI: 10.1038/nrmicro1532
Genomics meets HIV-1
Abstract
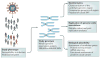
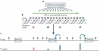
Genomics is now a core element in the effort to develop a vaccine against HIV-1. Thanks to unprecedented progress in high-throughput genotyping and sequencing, in knowledge about genetic variation in humans, and in evolutionary genomics, it is finally possible to systematically search the genome for common genetic variants that influence the human response to HIV-1. The identification of such variants would help to determine which aspects of the response to the virus are the most promising targets for intervention. However, a key obstacle to progress remains the scarcity of appropriate human cohorts available for genomic research.
Conflict of interest statement
Competing interests statement
The authors declare no competing financial interests.
Figures


References
-
- Ioannidis JP. Commentary: grading the credibility of molecular evidence for complex diseases. Int J Epidemiol. 2006;35:572–578. - PubMed
-
- O'Brien SJ, Nelson GW. Human genes that limit AIDS. Nature Genet. 2004;36:565–574. - PubMed
-
- Telenti A, Bleiber G. Host genetics of HIV-1 susceptibility. Future Virol. 2006;1:55–70.
-
- Todd JA. Statistical false positive or true disease pathway? Nature Genet. 2006;38:731–733. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

