Expression and loss of alleles in cultured mouse embryonic fibroblasts and stem cells carrying allelic fluorescent protein genes
- PMID: 17042952
- PMCID: PMC1621078
- DOI: 10.1186/1471-2199-7-36
Expression and loss of alleles in cultured mouse embryonic fibroblasts and stem cells carrying allelic fluorescent protein genes
Abstract
Background: Loss of heterozygosity (LOH) contributes to many cancers, but the rate at which these events occur in normal cells of the body is not clear. LOH would be detectable in diverse cell types in the body if this event were to confer an obvious cellular phenotype. Mice that carry two different fluorescent protein genes as alleles of a locus would seem to be a useful tool for addressing this issue because LOH would change a cell's phenotype from dichromatic to monochromatic. In addition, LOH caused by mitotic crossing over might be discernable in tissues because this event produces a pair of neighboring monochromatic cells that are different colors.
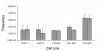
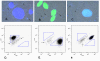
Results: As a step in assessing the utility of this approach, we derived primary embryonic fibroblast populations and embryonic stem cell lines from mice that carried two different fluorescent protein genes as alleles at the chromosome 6 locus, ROSA26. Fluorescence activated cell sorting (FACS) showed that the vast majority of cells in each line expressed the two marker proteins at similar levels, and that populations exhibited expression noise similar to that seen in bacteria and yeast. Cells with a monochromatic phenotype were present at frequencies on the order of 10(-4) and appeared to be produced at a rate of approximately 10(-5) variant cells per mitosis. 45 of 45 stably monochromatic ES cell clones exhibited loss of the expected allele at the ROSA26 locus. More than half of these clones retained heterozygosity at a locus between ROSA26 and the centromere. Other clones exhibited LOH near the centromere, but were disomic for chromosome 6.
Conclusion: Allelic fluorescent markers allowed LOH at the ROSA26 locus to be detected by FACS. LOH at this locus was usually not accompanied by LOH near the centromere, suggesting that mitotic recombination was the major cause of ROSA26 LOH. Dichromatic mouse embryonic cells provide a novel system for studying genetic/karyotypic stability and factors influencing expression from allelic genes. Similar approaches will allow these phenomena to be studied in tissues.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

