Effect of force on mononucleosomal dynamics
- PMID: 17043216
- PMCID: PMC1635095
- DOI: 10.1073/pnas.0607526103
Effect of force on mononucleosomal dynamics
Abstract
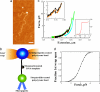
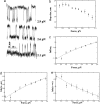
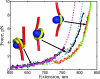
Using single-molecule optical-trapping techniques, we examined the force-induced dynamic behavior of a single nucleosome core particle. Our experiments using the DNA construct containing the 601 nucleosome-positioning sequence revealed that the nucleosome unravels in at least two major stages. The first stage, which we attributed to the unraveling of the first DNA wrap around the histone octamer, could be mechanically induced in a reversible manner, and when kept at constant force within a critical force range, exhibited two-state hopping behavior. From the hopping data, we determined the force-dependent equilibrium constant and rates for opening/closing of the outer wrap. Our investigation of the second unraveling event at various loading rates, which we attributed to the inner DNA wrap, revealed that this unraveling event cannot be described as a simple two-state process. We also looked at the behavior of the mononucleosome in a high-salt buffer, which revealed that the outer DNA wrap is more sensitive to changes in the ionic environment than the inner DNA wrap. These findings are needed to understand the energetics of nucleosome remodeling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

