The influence of recombination on human genetic diversity
- PMID: 17044736
- PMCID: PMC1575889
- DOI: 10.1371/journal.pgen.0020148
The influence of recombination on human genetic diversity
Abstract
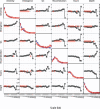
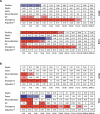
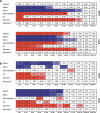
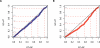
In humans, the rate of recombination, as measured on the megabase scale, is positively associated with the level of genetic variation, as measured at the genic scale. Despite considerable debate, it is not clear whether these factors are causally linked or, if they are, whether this is driven by the repeated action of adaptive evolution or molecular processes such as double-strand break formation and mismatch repair. We introduce three innovations to the analysis of recombination and diversity: fine-scale genetic maps estimated from genotype experiments that identify recombination hotspots at the kilobase scale, analysis of an entire human chromosome, and the use of wavelet techniques to identify correlations acting at different scales. We show that recombination influences genetic diversity only at the level of recombination hotspots. Hotspots are also associated with local increases in GC content and the relative frequency of GC-increasing mutations but have no effect on substitution rates. Broad-scale association between recombination and diversity is explained through covariance of both factors with base composition. To our knowledge, these results are the first evidence of a direct and local influence of recombination hotspots on genetic variation and the fate of individual mutations. However, that hotspots have no influence on substitution rates suggests that they are too ephemeral on an evolutionary time scale to have a strong influence on broader scale patterns of base composition and long-term molecular evolution.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures


References
-
- Maynard Smith J, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23:23–35. - PubMed
-
- Nachman MW. Single nucleotide polymorphisms and recombination rate in humans. Trends Genet. 2001;17:481–485. - PubMed
-
- Payseur BA, Nachman MW. Gene density and human nucleotide polymorphism. Mol Biol Evol. 2002;19:336–340. - PubMed
-
- Wolfe KH, Sharp PM, Li WH. Mutation rates differ among regions of the mammalian genome. Nature. 1989;337:283–285. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

