A baculovirus alkaline nuclease knockout construct produces fragmented DNA and aberrant capsids
- PMID: 17046043
- PMCID: PMC1852455
- DOI: 10.1016/j.virol.2006.09.008
A baculovirus alkaline nuclease knockout construct produces fragmented DNA and aberrant capsids
Abstract
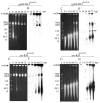
DNA replication of bacmid-derived constructs of the Autographa californica multiple nucleocapsid nucleopolyhedrovirus (AcMNPV) was analyzed by field inversion gel electrophoresis (FIGE) in combination with digestion at a unique Eco81I restriction enzyme site. Three constructs were characterized: a parental bacmid, a bacmid deleted for the alkaline nuclease gene, and a bacmid from which the gp64 gene had been deleted. The latter was employed as a control for comparison with the alkaline nuclease knockout because neither yields infectious virus and their replication is limited to the initially transfected cells. The major difference between DNA replicated by the different constructs was the presence in the alkaline nuclease knockout of high concentrations of relatively small, subgenome length DNA in preparations not treated with Eco81I. Furthermore, upon Eco81I digestion, the alkaline nuclease knockout bacmid also yielded substantially more subgenome size DNA than the other constructs. Electron microscopic examination of cells transfected with the alkaline nuclease knockout indicated that, in addition to a limited number of normal-appearing electron-dense nucleocapsids, numerous aberrant capsid-like structures were observed indicating a defect in nucleocapsid maturation or in a DNA processing step that is necessary for encapsidation. Because of the documented role of the baculovirus alkaline nuclease and its homologs from other viruses in homologous recombination, these data suggest that DNA recombination may play a major role in the production of baculovirus genomes.
Figures



Similar articles
-
Characterization of the role of very late expression factor 1 in baculovirus capsid structure and DNA processing.J Virol. 2006 Feb;80(4):1724-33. doi: 10.1128/JVI.80.4.1724-1733.2006. J Virol. 2006. PMID: 16439529 Free PMC article.
-
Autographa californica multiple nucleopolyhedrovirus nucleocapsid assembly is interrupted upon deletion of the 38K gene.J Virol. 2006 Dec;80(23):11475-85. doi: 10.1128/JVI.01155-06. Epub 2006 Sep 20. J Virol. 2006. PMID: 16987976 Free PMC article.
-
Characterization of a baculovirus lacking the alkaline nuclease gene.J Virol. 2004 Oct;78(19):10650-6. doi: 10.1128/JVI.78.19.10650-10656.2004. J Virol. 2004. PMID: 15367632 Free PMC article.
-
Nucleocapsid Assembly of Baculoviruses.Viruses. 2019 Jul 1;11(7):595. doi: 10.3390/v11070595. Viruses. 2019. PMID: 31266177 Free PMC article. Review.
-
Baculovirus Molecular Biology [Internet].3rd edition. Bethesda (MD): National Center for Biotechnology Information (US); 2013. 3rd edition. Bethesda (MD): National Center for Biotechnology Information (US); 2013. PMID: 24479205 Free Books & Documents. Review.
Cited by
-
The Autographa californica M nucleopolyhedrovirus ac79 gene encodes an early gene product with structural similarities to UvrC and intron-encoded endonucleases that is required for efficient budded virus production.J Virol. 2012 May;86(10):5614-25. doi: 10.1128/JVI.06252-11. Epub 2012 Mar 14. J Virol. 2012. PMID: 22419804 Free PMC article.
-
Characterization of a late gene, ORF75 from Bombyx mori nucleopolyhedrovirus.Mol Biol Rep. 2011 Mar;38(3):2141-9. doi: 10.1007/s11033-010-0341-6. Epub 2010 Sep 17. Mol Biol Rep. 2011. PMID: 20848211
-
Baculovirus infection induces a DNA damage response that is required for efficient viral replication.J Virol. 2011 Dec;85(23):12547-56. doi: 10.1128/JVI.05766-11. Epub 2011 Sep 14. J Virol. 2011. PMID: 21917957 Free PMC article.
-
Characterization of AcMNPV with a deletion of ac68 gene.Virus Genes. 2008 Aug;37(1):119-27. doi: 10.1007/s11262-008-0238-9. Epub 2008 May 16. Virus Genes. 2008. PMID: 18483845
-
Identification of Multiple Replication Stages and Origins in the Nucleopolyhedrovirus of Anticarsia gemmatalis.Viruses. 2019 Jul 15;11(7):648. doi: 10.3390/v11070648. Viruses. 2019. PMID: 31311127 Free PMC article.
References
-
- Anon . The MJ Research PPI-200 Programmable Power Inverter. 1993.
-
- Boehmer PE, Lehman IR. Herpes simplex virus DNA replication. Annu Rev Biochem. 1997;66:347–84. - PubMed
-
- Bujnicki J, Rychlewski L. The herpesvirus alkaline exonuclease belongs to the restriction endonuclease PD-(D/E)XK superfamily: insight from molecular modeling and phylogenetic analysis. Virus Genes. 2001;22(2):219–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

