Defects in energy homeostasis in Leigh syndrome French Canadian variant through PGC-1alpha/LRP130 complex
- PMID: 17050673
- PMCID: PMC1620022
- DOI: 10.1101/gad.1483906
Defects in energy homeostasis in Leigh syndrome French Canadian variant through PGC-1alpha/LRP130 complex
Abstract
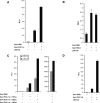
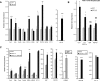
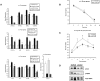
Leigh syndrome French Canadian variant (LSFC) is an autosomal recessive neurodegenerative disorder due to mutation in the LRP130 (leucine-rich protein 130 kDa) gene. Unlike classic Leigh syndrome, the French Canadian variant spares the heart, skeletal muscle, and kidneys, but severely affects the liver. The precise role of LRP130 in cytochrome c oxidase deficiency and hepatic lactic acidosis that accompanies this disorder is unknown. We show here that LRP130 is a component of the PGC-1alpha (peroxisome proliferator-activated receptor coactivator 1-alpha) transcriptional coactivator holocomplex and regulates expression of PEPCK (phosphoenolpyruvate carboxykinase), G6P (glucose-6-phosphatase), and certain mitochondrial genes through PGC-1alpha. Reduction of LRP130 in fasted mice via adenoviral RNA interference (RNAi) vector blocks the induction of PEPCK and G6P, and blunts hepatic glucose output. LRP130 is also necessary for PGC-1alpha-dependent transcription of several mitochondrial genes in vivo. These data link LRP130 and PGC-1alpha to defective hepatic energy homeostasis in LSFC, and reveal a novel regulatory mechanism of glucose homeostasis.
Figures




References
-
- Brunet, A., Bonni, A., Zigmond, M.J., Lin, M.Z., Juo, P., Hu, L.S., Anderson, M.J., Arden, K.C., Blenis, J., Greenberg, M.E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. - PubMed
-
- Erdjument-Bromage, H., Lui, M., Lacomis, L., Grewal, A., Annan, R.S., McNulty, D.E., Carr, S.A., Tempst, P. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J. Chromatogr. A. 1998;826:167–181. - PubMed
-
- Hall, R.K., Yamasaki, T., Kucera, T., Waltner-Law, M., O'Brien, R., Granner, D.K. Regulation of phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein-1 gene expression by insulin. The role of winged helix/forkhead proteins. J. Biol. Chem. 2000;275:30169–30175. - PubMed
-
- Herzig, S., Long, F., Jhala, U.S., Hedrick, S., Quinn, R., Bauer, A., Rudolph, D., Schutz, G., Yoon, C., Puigserver, P., et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. - PubMed
-
- Hou, J., Wang, F., McKeehan, W.L. Molecular cloning and expression of the gene for a major leucine-rich protein from human hepatoblastoma cells (HepG2) In Vitro Cell. Dev. Biol. Anim. 1994;30A:111–114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
