Microarray expression analysis of meiosis and microsporogenesis in hexaploid bread wheat
- PMID: 17052357
- PMCID: PMC1647286
- DOI: 10.1186/1471-2164-7-267
Microarray expression analysis of meiosis and microsporogenesis in hexaploid bread wheat
Abstract
Background: Our understanding of the mechanisms that govern the cellular process of meiosis is limited in higher plants with polyploid genomes. Bread wheat is an allohexaploid that behaves as a diploid during meiosis. Chromosome pairing is restricted to homologous chromosomes despite the presence of homoeologues in the nucleus. The importance of wheat as a crop and the extensive use of wild wheat relatives in breeding programs has prompted many years of cytogenetic and genetic research to develop an understanding of the control of chromosome pairing and recombination. The rapid advance of biochemical and molecular information on meiosis in model organisms such as yeast provides new opportunities to investigate the molecular basis of chromosome pairing control in wheat. However, building the link between the model and wheat requires points of data contact.
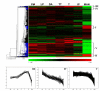
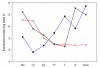
Results: We report here a large-scale transcriptomics study using the Affymetrix wheat GeneChip(R) aimed at providing this link between wheat and model systems and at identifying early meiotic genes. Analysis of the microarray data identified 1,350 transcripts temporally-regulated during the early stages of meiosis. Expression profiles with annotated transcript functions including chromatin condensation, synaptonemal complex formation, recombination and fertility were identified. From the 1,350 transcripts, 30 displayed at least an eight-fold expression change between and including pre-meiosis and telophase II, with more than 50% of these having no similarities to known sequences in NCBI and TIGR databases.
Conclusion: This resource is now available to support research into the molecular basis of pairing and recombination control in the complex polyploid, wheat.
Figures






References
-
- Francki M, Carter M, Ryan K, Hunter A, Bellgard M, Appels R. Comparative organization of wheat homoeologous group 3S and 7L using wheat-rice synteny and identification of potential markers for genes controlling xanthophyll content in wheat. Funct Integr Genom. 2004;4:118–130. doi: 10.1007/s10142-004-0110-5. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

