inGeno--an integrated genome and ortholog viewer for improved genome to genome comparisons
- PMID: 17054788
- PMCID: PMC1635569
- DOI: 10.1186/1471-2105-7-461
inGeno--an integrated genome and ortholog viewer for improved genome to genome comparisons
Abstract
Background: Systematic genome comparisons are an important tool to reveal gene functions, pathogenic features, metabolic pathways and genome evolution in the era of post-genomics. Furthermore, such comparisons provide important clues for vaccines and drug development. Existing genome comparison software often lacks accurate information on orthologs, the function of similar genes identified and genome-wide reports and lists on specific functions. All these features and further analyses are provided here in the context of a modular software tool "inGeno" written in Java with Biojava subroutines.
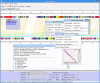
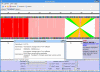
Results: InGeno provides a user-friendly interactive visualization platform for sequence comparisons (comprehensive reciprocal protein--protein comparisons) between complete genome sequences and all associated annotations and features. The comparison data can be acquired from several different sequence analysis programs in flexible formats. Automatic dot-plot analysis includes output reduction, filtering, ortholog testing and linear regression, followed by smart clustering (local collinear blocks; LCBs) to reveal similar genome regions. Further, the system provides genome alignment and visualization editor, collinear relationships and strain-specific islands. Specific annotations and functions are parsed, recognized, clustered, logically concatenated and visualized and summarized in reports.
Conclusion: As shown in this study, inGeno can be applied to study and compare in particular prokaryotic genomes against each other (gram positive and negative as well as close and more distantly related species) and has been proven to be sensitive and accurate. This modular software is user-friendly and easily accommodates new routines to meet specific user-defined requirements.
Figures

References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

