Gene expression signatures of morphologically normal breast tissue identify basal-like tumors
- PMID: 17054791
- PMCID: PMC1779486
- DOI: 10.1186/bcr1608
Gene expression signatures of morphologically normal breast tissue identify basal-like tumors
Abstract
Introduction: The role of the cellular microenvironment in breast tumorigenesis has become an important research area. However, little is known about gene expression in histologically normal tissue adjacent to breast tumor, if this is influenced by the tumor, and how this compares with non-tumor-bearing breast tissue.
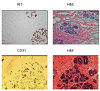
Methods: To address this, we have generated gene expression profiles of morphologically normal epithelial and stromal tissue, isolated using laser capture microdissection, from patients with breast cancer or undergoing breast reduction mammoplasty (n = 44).
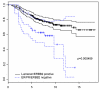
Results: Based on this data, we determined that morphologically normal epithelium and stroma exhibited distinct expression profiles, but molecular signatures that distinguished breast reduction tissue from tumor-adjacent normal tissue were absent. Stroma isolated from morphologically normal ducts adjacent to tumor tissue contained two distinct expression profiles that correlated with stromal cellularity, and shared similarities with soft tissue tumors with favorable outcome. Adjacent normal epithelium and stroma from breast cancer patients showed no significant association between expression profiles and standard clinical characteristics, but did cluster ER/PR/HER2-negative breast cancers with basal-like subtype expression profiles with poor prognosis.
Conclusion: Our data reveal that morphologically normal tissue adjacent to breast carcinomas has not undergone significant gene expression changes when compared to breast reduction tissue, and provide an important gene expression dataset for comparative studies of tumor expression profiles.
Figures








References
-
- Edwards BK, Brown ML, Wingo PA, Howe HL, Ward E, Ries LA, Schrag D, Jamison PM, Jemal A, Wu XC, et al. Annual report to the nation on the status of cancer, 1975–2002, featuring population-based trends in cancer treatment. J Natl Cancer Inst. 2005;97:1407–1427. - PubMed
-
- Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. - PubMed
-
- Torregrosa D, Bolufer P, Lluch A, Lopez JA, Barragan E, Ruiz A, Guillem V, Munarriz B, Garcia Conde J. Prognostic significance of c-erbB-2/neu amplification and epidermal growth factor receptor (EGFR) in primary breast cancer and their relation to estradiol receptor (ER) status. Clin Chim Acta. 1997;262:99–119. doi: 10.1016/S0009-8981(97)06542-X. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

