Intronic microRNA (miRNA)
- PMID: 17057362
- PMCID: PMC1559912
- DOI: 10.1155/JBB/2006/26818
Intronic microRNA (miRNA)
Abstract
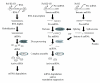
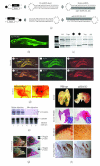
Nearly 97% of the human genome is composed of noncoding DNA, which varies from one species to another. Changes in these sequences often manifest themselves in clinical and circumstantial malfunction. Numerous genes in these non-protein-coding regions encode microRNAs, which are responsible for RNA-mediated gene silencing through RNA interference (RNAi)-like pathways. MicroRNAs (miRNAs), small single-stranded regulatory RNAs capable of interfering with intracellular messenger RNAs (mRNAs) with complete or partial complementarity, are useful for the design of new therapies against cancer polymorphisms and viral mutations. Currently, many varieties of miRNA are widely reported in plants, animals, and even microbes. Intron-derived microRNA (Id-miRNA) is a new class of miRNA derived from the processing of gene introns. The intronic miRNA requires type-II RNA polymerases (Pol-II) and spliceosomal components for their biogenesis. Several kinds of Id-miRNA have been identified in C elegans, mouse, and human cells; however, neither function nor application has been reported. Here, we show for the first time that intron-derived miRNAs are able to induce RNA interference in not only human and mouse cells, but in also zebrafish, chicken embryos, and adult mice, demonstrating the evolutionary preservation of intron-mediated gene silencing via functional miRNA in cell and in vivo. These findings suggest an intracellular miRNA-mediated gene regulatory system, fine-tuning the degradation of protein-coding messenger RNAs.
Figures







References
-
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Current Biology. 2003;13(10):807–818. - PubMed
-
- Lin SL, Chang D, Wu D-Y, Ying SY. A novel RNA splicing-mediated gene silencing mechanism potential for genome evolution. Biochemical and Biophysical Research Communications. 2003;310(3):754–760. - PubMed
-
- Lin SL, Chuong CM, Ying SY. A novel mRNA-cDNA interference phenomenon for silencing bcl-2 expression in human LNCaP cells. Biochemical and Biophysical Research Communications. 2001;281(3):639–644. - PubMed
-
- Ying SY, Lin SL. Intron-derived microRNAs - fine tuning of gene functions. Gene. 2004;342(1):25–28. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

