Generation of RNAi libraries for high-throughput screens
- PMID: 17057364
- PMCID: PMC1559919
- DOI: 10.1155/JBB/2006/45716
Generation of RNAi libraries for high-throughput screens
Abstract
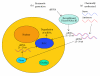
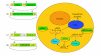
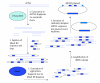
The completion of the genome sequencing for several organisms has created a great demand for genomic tools that can systematically analyze the growing wealth of data. In contrast to the classical reverse genetics approach of creating specific knockout cell lines or animals that is time-consuming and expensive, RNA-mediated interference (RNAi) has emerged as a fast, simple, and cost-effective technique for gene knockdown in large scale. Since its discovery as a gene silencing response to double-stranded RNA (dsRNA) with homology to endogenous genes in Caenorhabditis elegans (C elegans), RNAi technology has been adapted to various high-throughput screens (HTS) for genome-wide loss-of-function (LOF) analysis. Biochemical insights into the endogenous mechanism of RNAi have led to advances in RNAi methodology including RNAi molecule synthesis, delivery, and sequence design. In this article, we will briefly review these various RNAi library designs and discuss the benefits and drawbacks of each library strategy.
Figures
References
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. - PubMed
-
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123(4):607–620. - PubMed
-
- Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110(5):563–574. - PubMed
-
- Besse D, Budisa N, Karnbrock W, et al. Chalcogen-analogs of amino acids. Their use in X-ray crystallographic and folding studies of peptides and proteins. Biological Chemistry. 1997;378(3-4):211–218. - PubMed
LinkOut - more resources
Full Text Sources

