The crystal structure of the BAR domain from human Bin1/amphiphysin II and its implications for molecular recognition
- PMID: 17059209
- PMCID: PMC2572078
- DOI: 10.1021/bi060717k
The crystal structure of the BAR domain from human Bin1/amphiphysin II and its implications for molecular recognition
Abstract
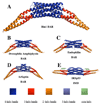
BAR domains are found in proteins that bind and remodel membranes and participate in cytoskeletal and nuclear processes. Here, we report the crystal structure of the BAR domain from the human Bin1 protein at 2.0 A resolution. Both the quaternary and tertiary architectures of the homodimeric Bin1BAR domain are built upon "knobs-into-holes" packing of side chains, like those found in conventional left-handed coiled-coils, and this packing governs the curvature of a putative membrane-engaging concave face. Our calculations indicate that the Bin1BAR domain contains two potential sites for protein-protein interactions on the convex face of the dimer. Comparative analysis of structural features reveals that at least three architectural subtypes of the BAR domain are encoded in the human genome, represented by the Arfaptin, Bin1/Amphiphysin, and IRSp53 BAR domains. We discuss how these principal groups may differ in their potential to form regulatory heterotypic interactions.
Figures






References
-
- Sakamuro D, Elliott KJ, Wechsler-Reya R, Prendergast GC. BIN1 is a novel MYC-interacting protein with features of a tumour suppressor. Nat Genet. 1996;14:69–77. - PubMed
-
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. - PubMed
-
- Tarricone C, Xiao B, Justin N, Walker PA, Rittinger K, Gamblin SJ, Smerdon SJ. The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways. Nature. 2001;411:215–219. - PubMed
-
- Weissenhorn W. Crystal structure of the endophilin-A1 BAR domain. J. Mol. Biol. 2005;351:653–661. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

