Meta-All: a system for managing metabolic pathway information
- PMID: 17059592
- PMCID: PMC1635068
- DOI: 10.1186/1471-2105-7-465
Meta-All: a system for managing metabolic pathway information
Abstract
Background: Many attempts are being made to understand biological subjects at a systems level. A major resource for these approaches are biological databases, storing manifold information about DNA, RNA and protein sequences including their functional and structural motifs, molecular markers, mRNA expression levels, metabolite concentrations, protein-protein interactions, phenotypic traits or taxonomic relationships. The use of these databases is often hampered by the fact that they are designed for special application areas and thus lack universality. Databases on metabolic pathways, which provide an increasingly important foundation for many analyses of biochemical processes at a systems level, are no exception from the rule. Data stored in central databases such as KEGG, BRENDA or SABIO-RK is often limited to read-only access. If experimentalists want to store their own data, possibly still under investigation, there are two possibilities. They can either develop their own information system for managing that own data, which is very time-consuming and costly, or they can try to store their data in existing systems, which is often restricted. Hence, an out-of-the-box information system for managing metabolic pathway data is needed.
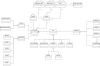
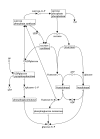
Results: We have designed META-ALL, an information system that allows the management of metabolic pathways, including reaction kinetics, detailed locations, environmental factors and taxonomic information. Data can be stored together with quality tags and in different parallel versions. META-ALL uses Oracle DBMS and Oracle Application Express. We provide the META-ALL information system for download and use. In this paper, we describe the database structure and give information about the tools for submitting and accessing the data. As a first application of META-ALL, we show how the information contained in a detailed kinetic model can be stored and accessed.
Conclusion: META-ALL is a system for managing information about metabolic pathways. It facilitates the handling of pathway-related data and is designed to help biochemists and molecular biologists in their daily research. It is available on the Web at http://bic-gh.de/meta-all and can be downloaded free of charge and installed locally.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

