ProFAT: a web-based tool for the functional annotation of protein sequences
- PMID: 17059594
- PMCID: PMC1636073
- DOI: 10.1186/1471-2105-7-466
ProFAT: a web-based tool for the functional annotation of protein sequences
Abstract
Background: The functional annotation of proteins relies on published information concerning their close and remote homologues in sequence databases. Evidence for remote sequence similarity can be further strengthened by a similar biological background of the query sequence and identified database sequences. However, few tools exist so far, that provide a means to include functional information in sequence database searches.
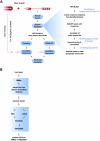
Results: We present ProFAT, a web-based tool for the functional annotation of protein sequences based on remote sequence similarity. ProFAT combines sensitive sequence database search methods and a fold recognition algorithm with a simple text-mining approach. ProFAT extracts identified hits based on their biological background by keyword-mining of annotations, features and most importantly, literature associated with a sequence entry. A user-provided keyword list enables the user to specifically search for weak, but biologically relevant homologues of an input query. The ProFAT server has been evaluated using the complete set of proteins from three different domain families, including their weak relatives and could correctly identify between 90% and 100% of all domain family members studied in this context. ProFAT has furthermore been applied to a variety of proteins from different cellular contexts and we provide evidence on how ProFAT can help in functional prediction of proteins based on remotely conserved proteins.
Conclusion: By employing sensitive database search programs as well as exploiting the functional information associated with database sequences, ProFAT can detect remote, but biologically relevant relationships between proteins and will assist researchers in the prediction of protein function based on remote homologies.
Figures





Similar articles
-
HMMerThread: detecting remote, functional conserved domains in entire genomes by combining relaxed sequence-database searches with fold recognition.PLoS One. 2011 Mar 10;6(3):e17568. doi: 10.1371/journal.pone.0017568. PLoS One. 2011. PMID: 21423752 Free PMC article.
-
Filling-in void and sparse regions in protein sequence space by protein-like artificial sequences enables remarkable enhancement in remote homology detection capability.J Mol Biol. 2014 Feb 20;426(4):962-79. doi: 10.1016/j.jmb.2013.11.026. Epub 2013 Dec 4. J Mol Biol. 2014. PMID: 24316367
-
WSsas: a web service for the annotation of functional residues through structural homologues.Bioinformatics. 2009 May 1;25(9):1192-4. doi: 10.1093/bioinformatics/btp116. Epub 2009 Feb 27. Bioinformatics. 2009. PMID: 19251774
-
Identifying remote protein homologs by network propagation.FEBS J. 2005 Oct;272(20):5119-28. doi: 10.1111/j.1742-4658.2005.04947.x. FEBS J. 2005. PMID: 16218946 Review.
-
Exploiting protein structure data to explore the evolution of protein function and biological complexity.Philos Trans R Soc Lond B Biol Sci. 2006 Mar 29;361(1467):425-40. doi: 10.1098/rstb.2005.1801. Philos Trans R Soc Lond B Biol Sci. 2006. PMID: 16524831 Free PMC article. Review.
Cited by
-
Cold-Induced Reprogramming of Subcutaneous White Adipose Tissue Assessed by Single-Cell and Single-Nucleus RNA Sequencing.Research (Wash D C). 2023 Jun 28;6:0182. doi: 10.34133/research.0182. eCollection 2023. Research (Wash D C). 2023. PMID: 37398933 Free PMC article.
-
Improving classification in protein structure databases using text mining.BMC Bioinformatics. 2009 May 5;10:129. doi: 10.1186/1471-2105-10-129. BMC Bioinformatics. 2009. PMID: 19416501 Free PMC article.
-
HMMerThread: detecting remote, functional conserved domains in entire genomes by combining relaxed sequence-database searches with fold recognition.PLoS One. 2011 Mar 10;6(3):e17568. doi: 10.1371/journal.pone.0017568. PLoS One. 2011. PMID: 21423752 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

