Analysis of Bartonella adhesin A expression reveals differences between various B. henselae strains
- PMID: 17060468
- PMCID: PMC1828432
- DOI: 10.1128/IAI.00963-06
Analysis of Bartonella adhesin A expression reveals differences between various B. henselae strains
Abstract
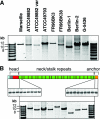
Bartonella henselae causes cat scratch disease and the vasculoproliferative disorders bacillary angiomatosis and peliosis hepatis in humans. One of the best known pathogenicity factors of B. henselae is Bartonella adhesin A (BadA), which is modularly constructed, consisting of head, neck/stalk, and membrane anchor domains. BadA is important for the adhesion of B. henselae to extracellular-matrix proteins and endothelial cells (ECs). In this study, we analyzed different B. henselae strains for BadA expression, autoagglutination, fibronectin (Fn) binding, and adhesion to ECs. We found that the B. henselae strains Marseille, ATCC 49882, Freiburg 96BK3 (FR96BK3), FR96BK38, and G-5436 express BadA. Remarkably, BadA expression was lacking in a B. henselae ATCC 49882 variant, in strains ATCC 49793 and Berlin-1, and in the majority of bacteria of strain Berlin-2. Adherence of B. henselae to ECs and Fn reliably correlated with BadA expression. badA was present in all tested strains, although the length of the gene varied significantly due to length variations of the stalk region. Sequencing of the promoter, head, and membrane anchor regions revealed only minor differences that did not correlate with BadA expression, apart from strain Berlin-1, in which a 1-bp deletion led to a frameshift in the head region of BadA. Our data suggest that, apart from the identified genetic modifications (frameshift deletion and recombination), other so-far-unknown regulatory mechanisms influence BadA expression. Because of variations between and within different B. henselae isolates, BadA expression should be analyzed before performing infection experiments with B. henselae.
Figures





References
-
- Alsmark, C. M., A. C. Frank, E. O. Karlberg, B. A. Legault, D. H. Ardell, B. Canback, A. S. Eriksson, A. K. Naslund, S. A. Handley, M. Huvet, B. LaScola, M. Holmberg, and S. G. Andersson. 2004. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc. Natl. Acad. Sci. USA 101:9716-9721. - PMC - PubMed
-
- Arvand, M., C. Wendt, T. Regnath, R. Ullrich, and H. Hahn. 1998. Characterization of Bartonella henselae isolated from bacillary angiomatosis lesions in a human immunodeficiency virus-infected patient in Germany. Clin. Infect. Dis. 26:1296-1299. - PubMed
-
- Centers for Disease Control and Prevention. 1999. Serodiagnosis of emerging infectious diseases: Bartonella and Ehrlichia infections. Centers for Disease Control and Prevention, Manassas, VA.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

