A role for Dicer in immune regulation
- PMID: 17060477
- PMCID: PMC2118134
- DOI: 10.1084/jem.20061692
A role for Dicer in immune regulation
Abstract
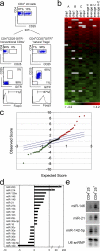
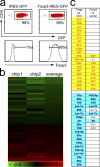
Micro RNAs (miRNAs) regulate gene expression at the posttranscriptional level. Here we show that regulatory T (T reg) cells have a miRNA profile distinct from conventional CD4 T cells. A partial T reg cell-like miRNA profile is conferred by the enforced expression of Foxp3 and, surprisingly, by the activation of conventional CD4 T cells. Depleting miRNAs by eliminating Dicer, the RNAse III enzyme that generates functional miRNAs, reduces T reg cell numbers and results in immune pathology. Dicer facilitates, in a cell-autonomous fashion, the development of T reg cells in the thymus and the efficient induction of Foxp3 by transforming growth factor beta. These results suggest that T reg cell development involves Dicer-generated RNAs.
Figures




References
-
- Fisher, A.G. 2002. Cellular identity and lineage choice. Nat. Rev. Immunol. 2:977–982. - PubMed
-
- Bartel, D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297. - PubMed
-
- Denli, A.M., B.B. Tops, R.H. Plasterk, R.F. Ketting, and G.J. Hannon. 2004. Processing of primary microRNAs by the Microprocessor complex. Nature. 432:231–235. - PubMed
-
- Gregory, R.I., K.P. Yan, G. Amuthan, T. Chendrimada, B. Doratotaj, N. Cooch, and R. Shiekhattar. 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature. 432:235–240. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

