Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1
- PMID: 17060499
- PMCID: PMC2064572
- DOI: 10.1083/jcb.200603149
Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1
Abstract
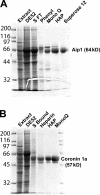
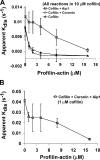
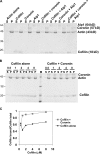
Actin filaments in cells depolymerize rapidly despite the presence of high concentrations of polymerizable G actin. Cofilin is recognized as a key regulator that promotes actin depolymerization. In this study, we show that although pure cofilin can disassemble Listeria monocytogenes actin comet tails, it cannot efficiently disassemble comet tails in the presence of polymerizable actin. Thymus extracts also rapidly disassemble comet tails, and this reaction is more efficient than pure cofilin when normalized to cofilin concentration. By biochemical fractionation, we identify Aip1 and coronin as two proteins present in thymus extract that facilitate the cofilin-mediated disassembly of Listeria comet tails. Together, coronin and Aip1 lower the amount of cofilin required to disassemble the comet tail and permit even low concentrations of cofilin to depolymerize actin in the presence of polymerizable G actin. The cooperative activities of cofilin, coronin, and Aip1 should provide a biochemical basis for understanding how actin filaments can grow in some places in the cell while shrinking in others.
Figures





References
-
- Balcer, H.I., A.L. Goodman, A.A. Rodal, E. Smith, J. Kugler, J.E. Heuser, and B.L. Goode. 2003. Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr. Biol. 13:2159–2169. - PubMed
-
- Bamburg, J.R. 1999. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15:185–230. - PubMed
-
- Cameron, L.A., T.M. Svitkina, D. Vignjevic, J.A. Theriot, and G.G. Borisy. 2001. Dendritic organization of actin comet tails. Curr. Biol. 11:130–135. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

