Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells
- PMID: 17060623
- PMCID: PMC1637589
- DOI: 10.1073/pnas.0605633103
Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells
Abstract
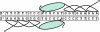
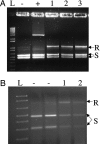
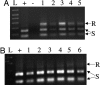
Zinc-finger nucleases are chimeric proteins consisting of engineered zinc-finger DNA-binding motifs attached to an endonuclease domain. These proteins can induce site-specific DNA double-strand breaks in genomic DNA, which are then substrates for cellular repair mechanisms. Here, we demonstrate that engineered zinc-finger nucleases function effectively in somatic cells of the nematode Caenorhabditis elegans. Although gene-conversion events were indistinguishable from uncut DNA in our assay, nonhomologous end joining resulted in mutations at the target site. A synthetic target on an extrachromosomal array was targeted with a previously characterized nuclease, and an endogenous genomic sequence was targeted with a pair of specifically designed nucleases. In both cases, approximately 20% of the target sites were mutated after induction of the corresponding nucleases. Alterations in the extrachromosomal targets were largely products of end-filling and blunt ligation. By contrast, alterations in the chromosomal target were mostly deletions. We interpret these differences to reflect the abundance of homologous templates present in the extrachromosomal arrays versus the paucity of such templates for repair of chromosomal breaks. In addition, we find evidence for the involvement of error-prone DNA synthesis in both homologous and nonhomologous pathways of repair. DNA ligase IV is required for efficient end joining, particularly of blunt ends. In its absence, a secondary end-joining pathway relies more heavily on microhomologies in producing deletions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

