Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model
- PMID: 17060627
- PMCID: PMC1637566
- DOI: 10.1073/pnas.0604817103
Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model
Abstract
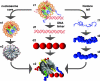
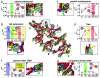
The role of each histone tail in regulating chromatin structure is elucidated by using a coarse-grained model of an oligonucleosome incorporating flexible histone tails that reproduces the conformational and dynamical properties of chromatin. Specifically, a tailored configurational-bias Monte Carlo method that efficiently samples the possible conformational states of oligonucleosomes yields positional distributions of histone tails around nucleosomes and illuminates the nature of tail/core/DNA interactions at various salt milieus. Analyses indicate that the H4 histone tails are most important in terms of mediating internucleosomal interactions, especially in highly compact chromatin with linker histones, followed by H3, H2A, and H2B tails in decreasing order of importance. In addition to mediating internucleosomal interactions, the H3 histone tails crucially screen the electrostatic repulsion between the entering/exiting DNA linkers. The H2A and H2B tails distribute themselves along the periphery of chromatin fibers and are important for mediating fiber/fiber interactions. A delicate balance between tail-mediated internucleosomal attraction and repulsion among linker DNAs allows the entering/exiting linker DNAs to align perpendicular to each other in linker-histone deficient chromatin, leading to the formation of an irregular zigzag-folded fiber with dominant pair-wise interactions between nucleosomes i and i +/- 4.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Felsenfeld G, Groudine M. Nature. 2003;421:448–453. - PubMed
-
- Richmond TJ, Davey CA. Nature. 2003;423:145–150. - PubMed
-
- Wade PA, Pruss D, Wolffe AP. Trends Bio Sci. 1997;22:128–132. - PubMed
-
- Davie JR. In: Chromatin Structure and Dynamics: State-of-the-Art. Zlatanova J, Leuba SH, editors. Amsterdam, The Netherlands: Elsevier; 2004. pp. 205–240.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

