Application of array-based comparative genomic hybridization to clinical diagnostics
- PMID: 17065418
- PMCID: PMC1876176
- DOI: 10.2353/jmoldx.2006.060029
Application of array-based comparative genomic hybridization to clinical diagnostics
Abstract
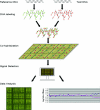
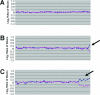
Microarray-based comparative genomic hybridization (array CGH) is a revolutionary platform that was recently adopted in the clinical laboratory. This technology was first developed as a research tool for the investigation of genomic alterations in cancer. It allows for a high-resolution evaluation of DNA copy number alterations associated with chromosome abnormalities. Array CGH is based on the use of differentially labeled test and reference genomic DNA samples that are simultaneously hybridized to DNA targets arrayed on a glass slide or other solid platform. In this review, we examine the technology and its transformation from a research tool into a maturing diagnostic instrument. We also evaluate the various approaches that have shaped the current platforms that are used for clinical applications. Finally, we discuss the advantages and shortcomings of "whole-genome" arrays and compare their diagnostic use to "targeted" arrays. Depending on their design, microarrays provide distinct advantages over conventional cytogenetic analysis because they have the potential to detect the majority of microscopic and submicroscopic chromosomal abnormalities. This new platform is poised to revolutionize modern cytogenetic diagnostics and to provide clinicians with a powerful tool to use in their increasingly sophisticated diagnostic capabilities.
Figures
Comment in
-
Diagnostic array comparative genomic hybridization: is it ready for prime time?J Mol Diagn. 2006 Nov;8(5):527. doi: 10.2353/jmoldx.2006.060152. J Mol Diagn. 2006. PMID: 17065417 Free PMC article. No abstract available.
-
Diagnostic genome profiling: unbiased whole genome or targeted analysis?J Mol Diagn. 2006 Nov;8(5):534-7; discussion 537-9. doi: 10.2353/jmoldx.2006.060131. J Mol Diagn. 2006. PMID: 17065419 Free PMC article. Review. No abstract available.
-
Whole-genome array comparative genome hybridization: the preferred diagnostic choice in postnatal clinical cytogenetics.J Mol Diagn. 2007 Apr;9(2):277. doi: 10.2353/jmoldx.2007.060187. J Mol Diagn. 2007. PMID: 17384222 Free PMC article. No abstract available.
References
-
- Lucito R, Healy J, Alexander J, Reiner A, Esposito D, Chi M, Rodgers L, Brady A, Sebat J, Troge J, West JA, Rostan S, Nguyen KC, Powers S, Ye KQ, Olshen A, Venkatraman E, Norton L, Wigler M. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2305. - PMC - PubMed
-
- Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. - PubMed
-
- Bejjani BA, Saleki R, Ballif BC, Rorem EA, Sundin K, Theisen A, Kashork CD, Shaffer LG. Use of targeted array-based CGH for the clinical diagnosis of chromosomal imbalance: Is less more? Am J Med Genet A. 2005;134:259–267. - PubMed
-
- Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, Chi B, Coe BP, Snijders A, Albertson DG, Pinkel D, Marra MA, Ling V, MacAulay C, Lam WL. A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet. 2004;36:299–303. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

