Evolution of the Yellow/Major Royal Jelly Protein family and the emergence of social behavior in honey bees
- PMID: 17065613
- PMCID: PMC1626640
- DOI: 10.1101/gr.5012006
Evolution of the Yellow/Major Royal Jelly Protein family and the emergence of social behavior in honey bees
Abstract
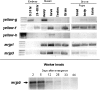
The genomic architecture underlying the evolution of insect social behavior is largely a mystery. Eusociality, defined by overlapping generations, parental brood care, and reproductive division of labor, has most commonly evolved in the Hymenopteran insects, including the honey bee Apis mellifera. In this species, the Major Royal Jelly Protein (MRJP) family is required for all major aspects of eusocial behavior. Here, using data obtained from the A. mellifera genome sequencing project, we demonstrate that the MRJP family is encoded by nine genes arranged in an approximately 60-kb tandem array. Furthermore, the MRJP protein family appears to have evolved from a single progenitor gene that encodes a member of the ancient Yellow protein family. Five genes encoding Yellow-family proteins flank the genomic region containing the genes encoding MRJPs. We describe the molecular evolution of these protein families. We then characterize developmental-stage-specific, sex-specific, and caste-specific expression patterns of the mrjp and yellow genes in the honey bee. We review empirical evidence concerning the functions of Yellow proteins in fruit flies and social ants, in order to shed light on the roles of both Yellow and MRJP proteins in A. mellifera. In total, the available evidence suggests that Yellows and MRJPs are multifunctional proteins with diverse, context-dependent physiological and developmental roles. However, many members of the Yellow/MRJP family act as facilitators of reproductive maturation. Finally, it appears that MRJP protein subfamily evolution from the Yellow protein family may have coincided with the evolution of honey bee eusociality.
Figures



References
-
- Albert S., Klaudiny J., Klaudiny J. The MRJP/YELLOW protein family of Apis mellifera: Identification of new members in the EST library. J. Insect Physiol. 2004;50:51–59. - PubMed
-
- Albert S., Bhattacharya D., Klaudiny J., Schmitzova J., Simuth J., Bhattacharya D., Klaudiny J., Schmitzova J., Simuth J., Klaudiny J., Schmitzova J., Simuth J., Schmitzova J., Simuth J., Simuth J. The family of major royal jelly proteins and its evolution. J. Mol. Evol. 1999a;49:290–297. - PubMed
-
- Albert S., Klaudiny J., Simuth J., Klaudiny J., Simuth J., Simuth J. Molecular characterization of MRJP3, highly polymorphic protein of honeybee (Apis mellifera) royal jelly. Insect Biochem. Mol. Biol. 1999b;29:427–434. - PubMed
-
- Albertova V., Su S., Brockmann A., Gadau J., Albert S., Su S., Brockmann A., Gadau J., Albert S., Brockmann A., Gadau J., Albert S., Gadau J., Albert S., Albert S. Organization and potential function of the mrjp3 locus in four honeybee species. J. Agric. Food Chem. 2005;53:8075–8081. - PubMed
-
- Artero R., Furlong E.E., Beckett K., Scott M.P., Baylies M., Furlong E.E., Beckett K., Scott M.P., Baylies M., Beckett K., Scott M.P., Baylies M., Scott M.P., Baylies M., Baylies M. Notch and Ras signaling pathway effector genes expressed in fusion competent and founder cells during Drosophila myogenesis. Development. 2003;130:6257–6272. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
